Next-generation sequencing (NGS) of DNA is routinely used in clinical testing. However, the complicated, multistep DNA extraction protocol and the unknown pre-analytic specimen handling often lead to sample-to-sample DNA contaminations. DNA contamination poses substantial challenges in NGS assays and affects diagnosis and treatment if contaminant DNA harbors clinically significant variants.
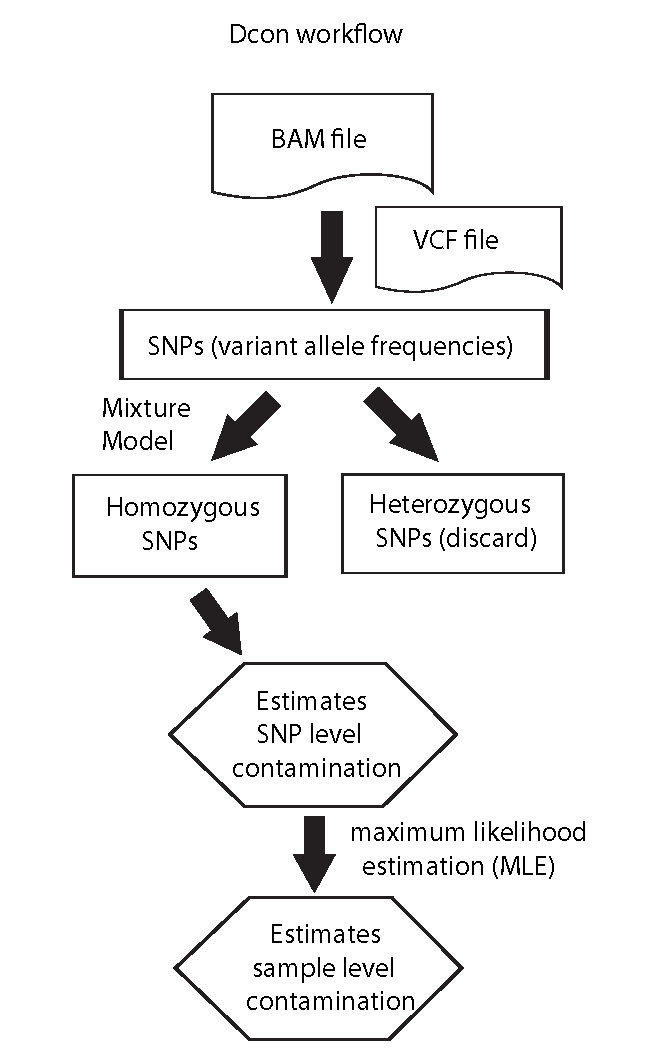
Dcon is a program to detect and quantify sample-to-sample germline DNA contaminations using the skewed (or shifted) distribution of variant allele frequency (VAF, VAF refers to the fraction of sequencing reads overlapping a genomic coordinate of a DNA variant such as SNP).
Unlike other tools, Dcon does NOT require any prior knowledge such as "genotype" and/or "allele frequencies in human population" calculated from 1000 Genome project. Dcon works on single BAM file and does not require tumor-normal pairs. Therefore, Dcon is also able to detect contamination from DNA sequencing data generated from non-human samples, where allele frequencies and genotype are generally not available.
Dcon requires python2.7 and these python packages (Python packages below will be automatically installed, please see instructions below).
Following instructions here to install pip. Please note: pip is already installed if you're using Python 2 >=2.7.9 or Python 3 >=3.4 binaries downloaded from python.org, but you'll need to upgrade pip.
Users have two ways to install Dcon:
-
Install Dcon from pypi:
- Open a terminal and type command:
pip install Dcon
- Open a terminal and type command:
-
Install Dcon from local directory
- Download Dcon directly from: this link
- If you download dcon-0.1.8.tar.gz. Go to the directory where dcon-0.1.8.tar.gz was saved. Open a terminal and type command:
tar -zxf dcon-0.1.8.tar.gz - If you download dcon-0.1.8.zip. Go to the directory where dcon-0.1.8.zip was saved. Open a terminal and type command:
unzip dcon-0.1.8.zip cd dcon-0.1.8python setup.py install# Use python2.7
User provides a BAM file and a VCF file. Variants in VCF file can be called from this particular BAM file or extracted from dbSNP (i.e. extract all SNPs that falling into captured genomic regions, Dcon automatically checks the VAF for each SNP and skips irrelevant SNPs).
python2.7 Dcon.py -b input.bam -v input.vcf -o output
Tips: How do I get a sub-section of a VCF file?
- Download and install tabix
- Using tabix to get a sub-section of a VCF file.
tabix -h ftp:https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20100804/ALL.2of4intersection.20100804.genotypes.vcf.gz 2:39967768-39967768 > output.vcf
General usage instruction is listed below:
Usage: Dcon.py [options]
Dcon: Estimate DNA Contamination from BAM file.
Options:
--version show program's version number and exit
-h, --help show this help message and exit
-b BAM_FILE, --bam=BAM_FILE
Alignment file in BAM format. BAM file must be indexed
and sorted using samTools
(https://samtools.sourceforge.net/).
-v VCF_FILE, --variant=VCF_FILE
VCF format file containing *candiate* SNPs (Note: Not
every SNP in this file has to be the real SNP in your
BAM file. They could be SNPs extracted from
dbSNP/1000Genome database, as long as they are located
in the captured region. However, too many spurious
SNPs will reduce the accuracy.). VCF file can be a
plain text, compressed (.gz, .z, .Z, .bz, .bz2 and
.gzip2) or remote file (https://, https://, ftp:https://).
Exclusive with '-r'.
-o OUTPUT_FILE, --output=OUTPUT_FILE
Prefix of output files. Will generate two intermediate
files: "prefix.SNP.tsv", "prefix.PI.filtered.tsv"
-m MIN_MAPQ, --mapq=MIN_MAPQ
Minimum mapping quality
(https://maq.sourceforge.net/qual.shtml). Mapping
quality is Phred-scaled probability of an alignment
being wrong. default=30
-q MIN_SEQQ, --seqq=MIN_SEQQ
Minimum base phred quality
(https://maq.sourceforge.net/qual.shtml). Base quality
is Phread-scaled probability of a base calling being
wrong. default=30
-c MIN_COVERAGE, --cvg=MIN_COVERAGE
Minimum number of reads supporting variant. default=30
-n PROCESSOR_NUM, --processor=PROCESSOR_NUM
Number of processes. default=1
-p PROBABILITY_CUT, --prob=PROBABILITY_CUT
Cutoff of probability. Probability is calcualted from
Bayesian Gaussian Mixture model to decicde if a SNP is
homozygous or heterozygous. default=0.5
-z ZSCORE_CUT, --zscore=ZSCORE_CUT
Cutoff of Z-score. The modified Z-score is calculated
from median absolute deviation. SNPs with Z-score
greater than this cutoff will be considered as outlier
and not used to estimate overall contamination level.
default=2.5
Targeted-capture sequencing data of 10 chronic myelomonocytic leukemia patients were downloaded from NCBI Sequence Read Archive (SRA).
- SRR6756023
- SRR6756025
- SRR6756028
- SRR6756033
- SRR6756036
- SRR6756040
- SRR6756042
- SRR6756045
- SRR6756048
- SRR6756053
A total of 45 synthetic datasets were created by mixing the above 10 original datasets in pairwise at 5%, 10%, ..., 45%, 50%. Mixed reads were then aligned to human genome (hg19) using BWA. Below are mixed BAM files from "SRR6756025" and "SRR6756028":
| File Name | Mix percentage | Reads from SRR6756025 | Reads from SRR6756028 | Total Reads | Download Links | MD5Sum of BAM file |
|---|---|---|---|---|---|---|
| SRR6756025_SRR6756028_P05.bam | 5% | 1,900,360 | 100,122 | 2,000,482 | bam, bai | 0fa86559117ee9f3070986593ff3b667 |
| SRR6756025_SRR6756028_P10.bam | 10% | 1,800,190 | 199,734 | 1,999,924 | bam, bai | 3e8705858900060836eccf984e4fe778 |
| SRR6756025_SRR6756028_P15.bam | 15% | 1,700,516 | 298,978 | 1,999,494 | bam, bai | 9ea836d52e7b78ec2e21d7fe8a547bdf |
| SRR6756025_SRR6756028_P20.bam | 20% | 1,600,992 | 399,158 | 2,000,150 | bam, bai | f2ed2b1cfbf89b6331f2a86065248767 |
| SRR6756025_SRR6756028_P25.bam | 25% | 1,501,164 | 499,074 | 2,000,238 | bam, bai | 3f838b28fa8898d12c6313de82473694 |
| SRR6756025_SRR6756028_P30.bam | 30% | 1,401,194 | 600,108 | 2,001,302 | bam, bai | 2b9392ef983e1e789b5d818c9f604b19 |
| SRR6756025_SRR6756028_P35.bam | 35% | 1,300,672 | 700,448 | 2,001,120 | bam, bai | 1d3a0fe4990bfa4da599df44d8f4c25a |
| SRR6756025_SRR6756028_P40.bam | 40% | 1,200,038 | 799,920 | 1,999,958 | bam, bai | e52bbeec296c2f933019a49e59639d46 |
| SRR6756025_SRR6756028_P45.bam | 45% | 1,099,964 | 899,424 | 1,999,388 | bam, bai | 72c8e3fca913735d1270875a3be59b17 |
| SRR6756025_SRR6756028_P50.bam | 50% | 999,998 | 998,942 | 1,998,940 | bam, bai | 75562d5c2ae4db37d2c97fe09c368222 |
- Download BAM files, BAM index files from the table above.
- Download the VCF file from here.
- Running Dcon to estimate contamination level.
#####################################
# Run Dcon on BAM file mixed at 5% #
#####################################
$ Dcon.py -v gene83exon.snp.vcf.gz -n 4 -b SRR6756025_SRR6756028_P05.bam -o P05
@ 2018-09-07 13:20:55: Running Dcon v0.1.8
@ 2018-09-07 13:20:55: Read SNPs ...
@ 2018-09-07 13:20:55: Estimating contamination for each variant. Saved to "P05.PI.tsv"
@ 2018-09-07 13:20:55: Building Bayesian Gaussian Mixture model (BGMM) ...
Converge status: True
Iterations: 3
@ 2018-09-07 13:20:55: Summerzie BGMM model ...
Means of homozygous component: 0.040164
Means of heterozygous component: 0.745609
Weight of homozygous component: 0.431413
Weight of heterozygous component: 0.568587
@ 2018-09-07 13:20:55: Classify variants ...
@ 2018-09-07 13:20:55: Writing to P05.PI.tsv ...
@ 2018-09-07 13:20:55: Filter outlier SNPs ...
@ 2018-09-07 13:20:55: Estimating overall contamination using maximum likelihood estimation (MLE) ...
@ 2018-09-07 13:20:55: Estimating contamination from homozygous SNPs ...
Overall contamination level of SRR6756025_SRR6756028_P05 is 0.051
#####################################
# Run Dcon on BAM file mixed at 10% #
#####################################
$ con.py -v gene83exon.snp.vcf.gz -n 4 -b SRR6756025_SRR6756028_P10.bam -o P10
@ 2018-09-07 13:23:08: Running Dcon v0.1.8
@ 2018-09-07 13:23:08: Read SNPs ...
@ 2018-09-07 13:23:08: Estimating contamination for each variant. Saved to "P10.PI.tsv"
@ 2018-09-07 13:23:08: Building Bayesian Gaussian Mixture model (BGMM) ...
Converge status: True
Iterations: 8
@ 2018-09-07 13:23:08: Summerzie BGMM model ...
Means of homozygous component: 0.067398
Means of heterozygous component: 0.702934
Weight of homozygous component: 0.421991
Weight of heterozygous component: 0.578009
@ 2018-09-07 13:23:08: Classify variants ...
@ 2018-09-07 13:23:08: Writing to P10.PI.tsv ...
@ 2018-09-07 13:23:08: Filter outlier SNPs ...
@ 2018-09-07 13:23:08: Estimating overall contamination using maximum likelihood estimation (MLE) ...
@ 2018-09-07 13:23:08: Estimating contamination from homozygous SNPs ...
Overall contamination level of SRR6756025_SRR6756028_P10 is 0.094
#####################################
# Run Dcon on BAM file mixed at 15% #
#####################################
$ Dcon.py -v gene83exon.snp.vcf.gz -n 4 -b SRR6756025_SRR6756028_P15.bam -o P15
@ 2018-09-07 13:23:51: Running Dcon v0.1.8
@ 2018-09-07 13:23:51: Read SNPs ...
@ 2018-09-07 13:23:51: Estimating contamination for each variant. Saved to "P15.PI.tsv"
@ 2018-09-07 13:23:51: Building Bayesian Gaussian Mixture model (BGMM) ...
Converge status: True
Iterations: 19
@ 2018-09-07 13:23:51: Summerzie BGMM model ...
Means of homozygous component: 0.107931
Means of heterozygous component: 0.687673
Weight of homozygous component: 0.420852
Weight of heterozygous component: 0.579148
@ 2018-09-07 13:23:51: Classify variants ...
@ 2018-09-07 13:23:51: Writing to P15.PI.tsv ...
@ 2018-09-07 13:23:51: Filter outlier SNPs ...
@ 2018-09-07 13:23:51: Estimating overall contamination using maximum likelihood estimation (MLE) ...
@ 2018-09-07 13:23:51: Estimating contamination from homozygous SNPs ...
Overall contamination level of SRR6756025_SRR6756028_P15 is 0.145
#####################################
# Run Dcon on BAM file mixed at 20% #
#####################################
$ Dcon.py -v gene83exon.snp.vcf.gz -n 4 -b SRR6756025_SRR6756028_P20.bam -o P20
@ 2018-09-07 13:24:17: Running Dcon v0.1.8
@ 2018-09-07 13:24:17: Read SNPs ...
@ 2018-09-07 13:24:17: Estimating contamination for each variant. Saved to "P20.PI.tsv"
@ 2018-09-07 13:24:17: Building Bayesian Gaussian Mixture model (BGMM) ...
Converge status: True
Iterations: 21
@ 2018-09-07 13:24:17: Summerzie BGMM model ...
Means of homozygous component: 0.137115
Means of heterozygous component: 0.657750
Weight of homozygous component: 0.417032
Weight of heterozygous component: 0.582968
@ 2018-09-07 13:24:17: Classify variants ...
@ 2018-09-07 13:24:17: Writing to P20.PI.tsv ...
@ 2018-09-07 13:24:17: Filter outlier SNPs ...
@ 2018-09-07 13:24:17: Estimating overall contamination using maximum likelihood estimation (MLE) ...
@ 2018-09-07 13:24:17: Estimating contamination from homozygous SNPs ...
Overall contamination level of SRR6756025_SRR6756028_P20 is 0.198
Dcon will generate 2 intermediate files
- prefix.SNP.tsv
- column-1: Chromosome ID
- column-2: SNP Position
- column-3: Allele 1
- column-4: Number of reads supporting Allele 1
- column-5: Allele 2
- column-6: Number of reads supporting Allele 1
- prefix.PI.filtered.tsv
- column-1: Chromosome ID
- column-2: SNP Position
- column-3: Allele 1
- column-4: Number of reads supporting Allele 1
- column-5: Allele 2
- column-6: Number of reads supporting Allele 2
- column-7: Ratio. {Number of reads supporting less frequent allele}/{Total reads covering this SNP}
- column-8: Probability of homozygous SNP
- column-9: Probability of heterozygous SNP
- column-10: Label. "Hom" or "Het"
- column-11: Distance used to rank SNP. Contamination calculated from SNPs with smaller distance is more reliable.
- column-12: Contamination percentage calculated from this individual SNP.
- column-13: Indicator of whether this SNP has passed outlier filtering.
Example of prefix.SNP.tsv
$ head P05.SNP.tsv
1 36937059 A 126 G 117
1 36937065 A 127 G 124
2 25469502 T 116 C 95
3 105439026 G 477 A 11
3 128199380 G 220 A 14
3 128199662 G 170 A 11
3 128204951 C 97 T 90
3 128205860 C 30 G 15
3 128206618 C 36 A 7
3 136056033 G 671 A 5
...
Example of prefix.PI.filtered.tsv
$ head P05.PI.tsv
Chrom Ref_pos Allele_1 Allele_1_count Allele_2 Allele_2_count Ratio Prob_of_Het Prob_of_Hom Label Distance Contamination_p Outlier
3 105439026 G 477 A 11 0.0230607966457 0.000182230455487 0.999817769545 Hom 0.0171029321787 0.0450819672131 Pass
3 128199380 G 220 A 14 0.0636363636364 0.000446156678585 0.999553843321 Hom 0.023472634812 0.119658119658 Pass
3 128199662 G 170 A 11 0.0647058823529 0.000458638756417 0.999541361244 Hom 0.0245421535286 0.121546961326 Pass
3 128206618 C 36 A 7 0.194444444444 0.0565034056397 0.94349659436 Hom 0.15428071562 0.325581395349 Fail
3 136056184 A 638 G 11 0.0172413793103 0.000164195828332 0.999835804172 Hom 0.022922349514 0.0338983050847 Pass
3 136088038 G 549 A 9 0.016393442623 0.000161803440411 0.99983819656 Hom 0.0237702862014 0.0322580645161 Pass
3 168801495 C 576 A 12 0.0208333333333 0.000174978971172 0.999825021029 Hom 0.019330395491 0.0408163265306 Pass
3 168801916 C 349 T 4 0.0114613180516 0.000148943307201 0.999851056693 Hom 0.0287024107728 0.0226628895184 Pass
3 168802737 G 278 A 3 0.0107913669065 0.000147326774526 0.999852673225 Hom 0.0293723619179 0.0213523131673 Pass
CPU model
Intel(R) Core(TM) i7-3720QM CPU @ 2.60GHz
Speed:
When VCF (9265 candidate SNPs within captured regions) file was provided , Dcon took 6 minutes and 5 seconds to estimate DNA contamination from a 177 Mb BAM file containing 2 million alignments. Using multiple threads (-p) will increase speed significantly.
In below figure, each boxplot is generated from 100 BAM files (Capture sequencing target 37 genes). And each BAM file is purposely mixed from two different samples at pre-defined percentages (10%, 20%, 30%, 40%, 50%). For example, in the leftmost boxplot(green), 10% of reads were from one sample, and 90% of reads were from another sample.
- Liguo Wang [email protected]
- Tao Ma [email protected]
- Rohan Gnanaolivu [email protected]
- Jagadheshwar Balan [email protected]
- Eric Klee Klee [email protected]
- Jean-Pierre Kocher [email protected]
This project is licensed under the GPL License - see the LICENSE file for details
This project is partially supported by the Clinical Genome Sequencing Laboratory (CGSL), Mayo Clinic.