Unsupervised outlier detection with privileged information. Generalizes one-class nu-SVM that estimates the support of a high-dimensional distribution by accounting for additional (privileged) set of features available in the training phase but not available in evaluating and predicting new data (for example, future time series behavior).
Along with the current SVM models, it solves dual optimization problem with respect to unknown coefficients with Sequential Minimal Optimization. SMO finds the two coefficients that violate Karush-Kuhn-Tucker conditions the most, and shifts them in opposite directions keeping their sum constant. The optimization procedure converges to a tau -approximate solution, which means that the KKT conditions are satisfied with the tolerance of tau.
You can just download the repository to your folder. Then from its root directory (where setup.py is located) run on the command line
python setup.py build_ext --inplace
After that, you can import the library from Python:
>>> import ocsvm_plus
Make sure you are using Python 3.x (you may need "python3" instead of "python"). You will also need Cython installed (pip install Cython or similar). CVXOPT is needed for comparison (pip install cvxopt). To be more confident, you should run unit tests:
python setup_debug.py build_ext --inplace
python -m unittest tests
Now debug version is also available:
>>> import ocsvm_plus_debug
The debug version performs assertions, obtains intermediate results in different ways and checks their equivalence, dumps more detailed info to the log file. It works much slower than basic (release) version.
class ocsvm_plus.OneClassSVM_plus(n_features, kernel='rbf', kernel_gamma='scale',
kernel_star='rbf', kernel_star_gamma='scale',
nu=0.5, gamma='auto', tau=0.001,
alg='best_step', ff_caches='not_bound',
kernel_cache_size=0, distance_cache_size=0,
max_iter=-1, random_seed=None,
logging_file_name=None)| Parameters: | Description |
|---|---|
| n_features: int | Number of original features. |
| kernel: {'rbf', 'linear'} or a class derived from ocsvm_plus.kernel, default='rbf' | Kernel K for original features X. |
| kernel_gamma: {'scale', 'auto'} or float, default='scale' | Kernel coefficient if K is 'rbf'. For kernel_gamma='scale' (default) it uses kernel_gamma=1/(n_features*X.var()) as value of kernel_gamma. For 'auto' it uses kernel_gamma=1/n_features. |
| kernel_star: {'rbf', 'linear'} or a class derived from ocsvm_plus.kernel, default='rbf' | Kernel K* for privileged features X_star. |
| kernel_star_gamma: {'scale', 'auto'} or float, default='scale' | Kernel coefficient if K* is 'rbf'. For kernel_star_gamma='scale' (default) it uses kernel_star_gamma=1/(n_features_star*X_star.var()) as value of kernel_star_gamma. For 'auto' it uses kernel_star_gamma=1/n_features_star. |
| nu: float, default=0.5 | Parameter of nu-SVM, original features regularizer, should be between (0, 1). |
| gamma: 'auto' or float, default='auto' | Privileged features regularizer. For gamma='auto' it uses gamma=nu*n_samples. |
| tau: float, default=0.001 | Tolerance for stopping criterion. |
| alg: {'best_step_2d', 'best_step', 'delta_pair', 'cvxopt'}, default='best_step' | Mode for optimization procedure, affects processing time. For 'delta_pair' the algorithm tries to find the most delta-violating pair of privileged coefficients and, if no delta-pairs remained, then it looks for alpha-violating pair of original coefficients. Although delta-optimization step for a given delta-pair is cheaper in time than alpha-step for a pair of alpha-coefficients (due to decision and correcting function updates), this option usually demonstrates slow convergence. 'best_step' tries to find both the most (worst) alpha- and the most delta-violating pairs and the most violating one is selected for the optimization step. 'best_step_2d' is similar to 'best_step', but if the selected pair is also a violating pair according to another criterion (not necessarily being the most violating), then the two-dimensional optimization is applied (for example, if the most violating pair is alpha-pair of examples ij and delta KKT condition is also violated, then the shift for two coefficients alpha_ij and a shift for two coefficients delta_ij are found together). 'best_step_2d' and 'best_step' may demonstrate comparable fastness, because the optimization of two pairs of coefficients at once is at the same time more computationally expensive. cvxopt uses the general-purpose quadratic optimization solver CVXOPT, based on interior-point methods, which are less memory and time efficient compared to SMO. Added for comparison tests. |
| ff_caches: {'all', 'not_bound', 'not_zero'}, default='not_bound' | Sets ranges for caches of indices of training examples C and C* for which the values of decision f(x_i) and correcting f*(x*_i) functions (without their intercepts –rho and b*) are kept actual on every iteration using simple updates (by subtract old pair of coefficients and add new ones) and without need to full recalculation (summing over all non-zero coefficients). Affects processing time of the optimization procedure. alpha-pair is selected over C, delta-pair is over C*. A wider cache provides a better selection of violation pairs and less costly function recalculations, while a smaller cache results in faster search for violation pairs and fewer updated f (x_i) and f * (x *_i). 'all': both caches include all training examples. 'not_bound': C includes elements i for which alpha_i>0 or 0<delta_i<1, C* includes all elements. 'not_zero': C and C* covers all elements except those who has both alpha_i and delta_i zero. Generally, ff_cache is a dict with keys 'anot0_dnot01', 'anot0_d0', 'anot0_d1', 'a0_dnot01', 'a0_d1', 'a0_d0', corresponding to six subsets of elements (respectively, alpha_i>0, 0<delta_i<1; alpha_i>0, delta_i=0; alpha_i>0, delta_i=1; alpha_i=0, 0<delta_i<1; alpha_i=0, delta_i=1 and alpha_i=0, delta_i=0) and key values: 2 – a subset belongs to both C and C*, or 1 - belongs only to C*, or 0 - not in caches ('all' is equivalent to all key values are 2, 'not_zero' - all key values are 2 except ff_cache['a0_d0']=0, 'not_bound' is equivalent to ff_caches = {'anot0_dnot01': 2, 'anot0_d0': 2, 'anot0_d1': 2, 'a0_dnot01': 2, 'a0_d1': 1, 'a0_d0': 1}). |
| kernel_cache_size: int, default=0 | Size of a cache (number of elements) to store K and K* values according to LRU policy. If 0, then the kernels K(x_i, x_j) and K*(x*_i, x*_j) are calculated only once and stored as ij-elements of triangular matrices. Limited cache size is memory-efficient, while 0 is the most time-efficient setting. |
| distance_cache_size: int, default=0 | Size of a cache (number of elements) to store values (K_ii-2K_ij+K_jj)/(nu * n_samples) and (K*_ii-2K*_ij+K*_jj)/gamma, LRU policy is used. If 0, then once a value is calculated, it is stored in triangular matrix. Limited cache size is memory-efficient, while 0 is the most time-efficient setting. |
| max_iter: int, default=-1 | Hard limit on iterations within solver, or -1 for no limit. |
| random_seed: int or None, default=None | Random generator initialization for test repeatability. |
| logging_file_name: str, 'uuid', 'time' or None, default=None | Text file name to dump intermediate results of iterative process of model training. If None, then no logging is performed. For 'uuid' the filename is set to an universally unique identifier, for example, 592541c44ae044b3a3f24119b1951e63.log. For 'time' it is set to current time, i.e. YYYY-mm-dd-HH-MM-SS-ffffff.log. |
| Attributes: | Description |
|---|---|
| alphas_ | Dual coefficients alpha_i. |
| deltas_ | Dual coefficients delta_i. |
| rho_ | Decision function intercept. |
| b_star_ | Correcting function intercept. |
| alpha_support_ | Indices i of training examples such that alpha_i>0. |
| delta_support_ | Indices i of training examples such that 0<delta_i<1. |
| fit_status_ | Returns True if there were at least one non-bound coefficient 0<delta_k<1 to correctly find intercepts rho and b_star. Otherwise, b_star=-(f^*(x_m^*)+f^*(x_n^*))/2, where mn is delta-pair. In this case fit_status_ is set to False, and one should make tau smaller and/or increase max_iter. |
| Methods: | Description |
|---|---|
fit(Xall[, y=None]) |
Detects the soft boundary of the set of original vectors X[n_samples, n_features] accounting for privileged vectors X_star[n_samples, n_features_star], where Xall=[n_samples, n_features + n_features_star] comprises X in the first n_features columns and X_star in the last n_features_star columns. Xall is array-like, if not C-ordered contiguous array it is copied, y is ignored. Returns self. |
decision_function(X) |
Signed distance to the separating hyperplane in original feature space, positive for an inlier and negative for an outlier points. X is array-like, if not C-ordered contiguous array it is copied. First n_features columns are used, other columns are ignored. Returns ndarray of shape (X.shape[0], ). |
correcting_function(X_star) |
Slack variables modelled using privileged features. X_star is array-like, if not C-ordered contiguous array it is copied. Last n_features_star columns are used as privileged vectors, other columns are ignored. Returns ndarray of shape (X_star.shape[0], ). |
predict(X) |
Perform classification on samples in X. Similar to decision_function(X), but the output is +1 if the decision function is positive and -1 otherwise. |
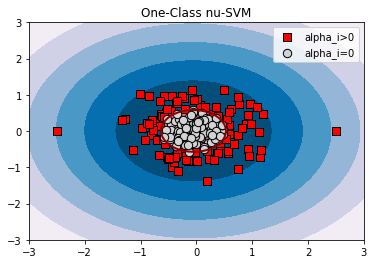
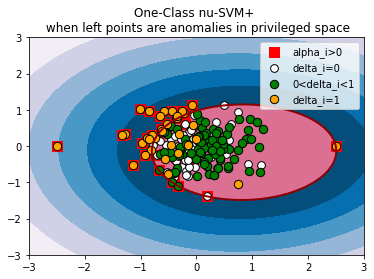
xx, yy = np.meshgrid(np.linspace(-3, 3, 500), np.linspace(-3, 3, 500))
np.random.seed(0)
X = 0.5*np.random.randn(200, 2)
X[-2:, :] = [[-2.5, 0], [2.5, 0]] # set left and right outliers
# OCSVM
ocsvm = OneClassSVM(nu=0.5, gamma=0.1).fit(X)
# OCSVM+
left = X[:, 0] < 0
X_star = np.empty((X.shape[0], 1))
X_star[~left] = 0.1*np.random.randn(X[~left].shape[0], 1) # right points spaced closely in privileged space
X_star[left] = (2*np.random.rand(X[left].shape[0], 1)-1) * 100
X_star[left] = X_star[left] + 10*np.sign(X_star[left]) # left points spaced chaotically in privileged space, (-110, -10) or (10, 110)
ocsvm_plus = OneClassSVM_plus(n_features=2, nu=0.5, gamma=0.1, kernel_gamma=0.1, kernel_star_gamma=0.1, tau=1e-3).fit(np.hstack((X, X_star)))
for title, model in [('One-Class nu-SVM', ocsvm),
('One-Class nu-SVM+ \n when left points are anomalies in privileged space', ocsvm_plus)]:
plt.figure()
plt.gca().set_title(title)
Z = model.decision_function(np.c_[xx.ravel(), yy.ravel()]).reshape(xx.shape)
plt.contourf(xx, yy, Z, levels=np.linspace(Z.min(), 0, 7), cmap=plt.cm.PuBu)
plt.contourf(xx, yy, Z, levels=[0, Z.max()], colors='palevioletred')
plt.contour(xx, yy, Z, levels=[0], linewidths=2, colors='darkred')
if 'One-Class nu-SVM+' in title:
if model.fit_status_:
plt.scatter(X[model.alpha_support_, 0], X[model.alpha_support_, 1], c='r', s=100, edgecolors='r', label='alpha_i>0', marker='s')
delta0 = [i for i in range(X.shape[0]) if model.deltas_[i] < 1e-05 and i not in model.delta_support_]
plt.scatter(X[delta0, 0], X[delta0, 1], c='w', s=60, edgecolors='k', label='delta_i=0')
plt.scatter(X[model.delta_support_, 0], X[model.delta_support_, 1], c='g', s=70, edgecolors='k', label='0<delta_i<1')
delta1 = [i for i in range(X.shape[0]) if model.deltas_[i] > 1-1e-05 and i not in model.delta_support_]
plt.scatter(X[delta1, 0], X[delta1, 1], c='orange', s=70, edgecolors='k', label='delta_i=1')
else: # OCSVM
plt.scatter(X[model.support_, 0], X[model.support_, 1], c='r', s=80, edgecolors='k', label='alpha_i>0', marker='s')
alpha0 = [i for i in range(X.shape[0]) if i not in model.support_]
plt.scatter(X[alpha0, 0], X[alpha0, 1], c='lightgrey', s=70, edgecolors='k', label='alpha_i=0')
plt.gca().legend()
OCSVM+ models the domain boundary distances (slack variables xi in the original nu-SVM, which characterize the measure of data point anomality) through parameterization with privileged features. In this example, the points to the left of the origin have a chaotic priviliged coordinate, while the points to the right are close in priviliged space. Such privileged information shifts approximated domain to the right, because the left points show higher anomality. For OCSVM the parameter nu=0.5 is both the upper bound of the fraction of anomalies and the lower bound of the fraction of support vectors. For OCSVM+ nu is still the upper bound of the fraction of anomalies, but no longer the lower bound of the fraction of support vectors.
STLCACHE library https://github.com/akashihi/stlcache is used for caching the values of kernel functions, many thanks to the authors.