Kyle Coleman*, Daiwei Zhang, Amelia Schroeder, Melanie Braisted, Niklas Blank, Alexis Jazmyn, Hanying Yan, Yanxiang Deng, Elizabeth F. Furth, Edward B. Lee, Christoph A. Thaiss, Jian Hu*, Mingyao Li*
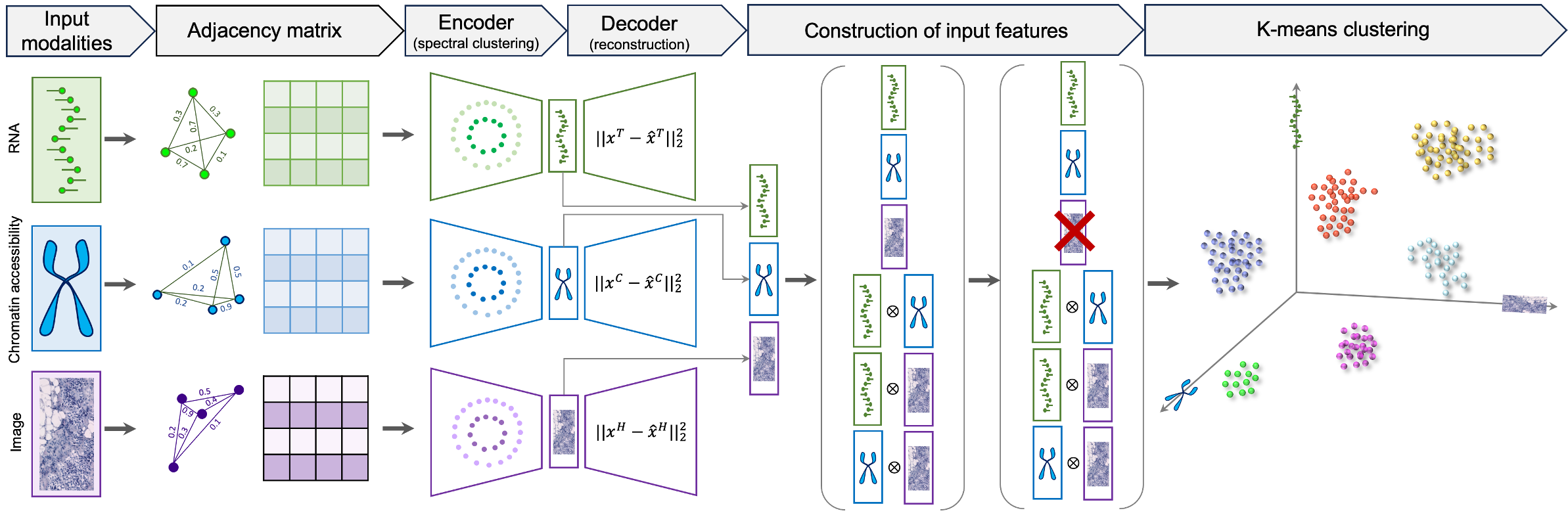
MISO is a deep-learning based method developed for the integration and clustering of multi-modal spatial omics data. MISO requires minimal hyperparameter tuning, and can be applied to any number of omic and imaging data modalities from any multi-modal spatial omics experiment. MISO has been evaluated on datasets from experiements including spatial transcriptomics (transcriptomics and histology), spatial epigenome-transcriptome co-profiling (chromatin accessibility, histone modification, and transcriptomics), spatial CITE-seq (transcriptomics, proteomics, and histology), and spatial transcriptomics and metabolomics (transcriptomics, metabolomics, and histology)
Typical install time is ~1 min.
MISO has been tested on the following operating systems:
- macOS: Ventura (13.5.1)
- Linux: CentOS (7)
MISO installation requires python version 3.7. The version of python can be checked by:
import platform
platform.python_version()'3.7.13'
We recommend creating and activating a new conda environment when installing the MISO package. For instance,
conda create -n miso python=3.7.13
conda activate misoThe MISO repository can be downloaded using:
git clone https://github.com/kpcoleman/misoThe pretrained ViT weights are stored on Git LFS, and can be downloaded using:
cd miso
git lfs install
git lfs fetch
git lfs pullThe MISO package and dependencies can then be installed:
python -m pip install .Typical runtime for MISO is ~1 min on a GPU.
For a tutorial, please see: https://github.com/kpcoleman/miso/blob/main/tutorial/tutorial.ipynb
The miso conda environment can be used for the tutorial by:
python -m pip install ipykernel
python -m ipykernel install --user --name=misoeinops==0.6.0
importlib
importlib-metadata
numpy==1.21.6
opencv_python==4.6.0.66
Pillow>=6.1.0
scanpy==1.9.1
scikit_image==0.19.3
scikit_learn==1.0.2
scipy==1.7.3
setuptools==65.6.3
torch==1.13.1
torchvision==0.14.1
tqdm==4.64.1
H&E image feature extraction code is based on HIPT and iSTAR. Pre-trained vision transformer models are from HIPT.