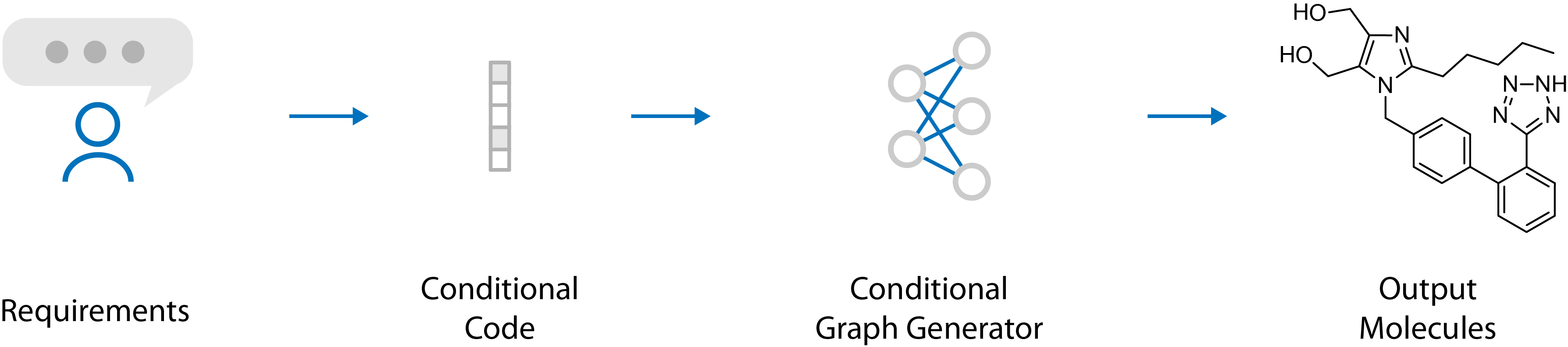
This repository contains the source code and data sets for the graph based molecule generator discussed in the article "Multi-Objective De Novo Drug Design with Conditional Graph Generative Model" (https://arxiv.org/abs/1801.07299).
Briefly speaking, we used conditional graph convolution to structure the generative model. The properties of output molecules can then be controlled using the conditional code.
This repo is built using Python 2.7, and utilizes the following packages:
- MXNet == 1.3.1
- RDKit == 2018.03.3
- Numpy
- Scikit-learn (for the predictive model)
To ease the installation process, please use the dockerfile environment defined in the Dockerfile.
train.py: main training script.mx_mg: package for the molecule generative model:data: packages for data processing workflows:conditionals.py: callables used to generate the conditional codes for moleculesdata_struct.py: defines atom types and bond typesdataloaders.py,datasets.pyandsamplers.py: data loading logicsutils.py: utility functions
models: library for graph generative modelsmodules.py: define modules (or blocks) such as graph convolutionnetworks.py: define networks (MolMP, MolRNN and CMolRNN)functions.py: autograd.Function objects and operations
builders.py: utilities for building molecules using generative models
rdkit_contrib: functions used to calculate QED and SAscore (for older version of rdkit)example.ipynb: tutorial
To train the model, first unpackdatasets.tar.gz (download here) to the current directory, and call:
./train.py {molmp|molrnn|scaffold|prop|kinase} path/to/outputWhere {molmp|molrnn|scaffold|prop|kinase} are model types, and path/to/output is the directory where you want to save the model's checkpoint file and log files. The following call:
./train.py {molmp|molrnn|scaffold|prop|kinase} -hgives help for each model type.
Please contact me. Email: [email protected] or [email protected]