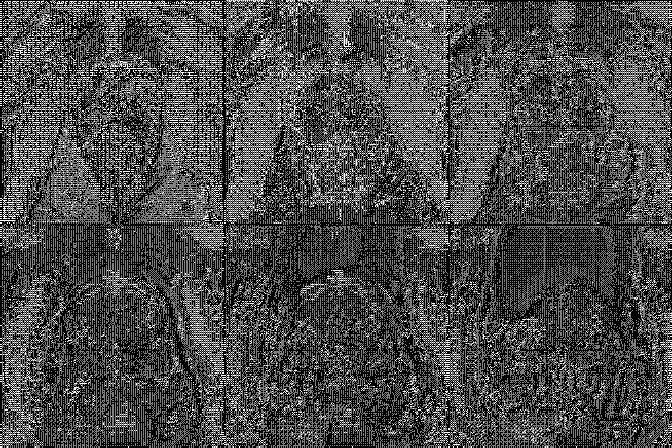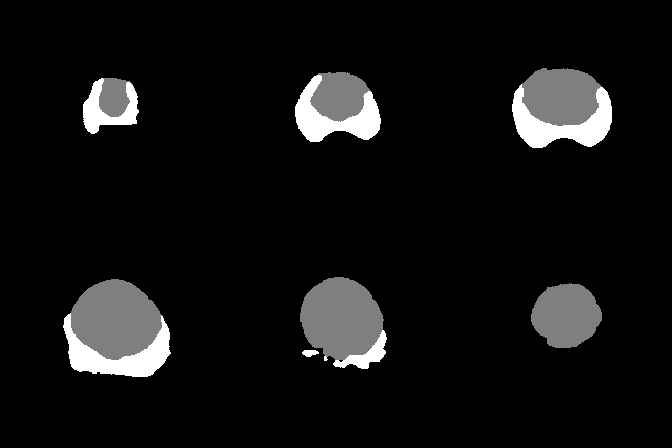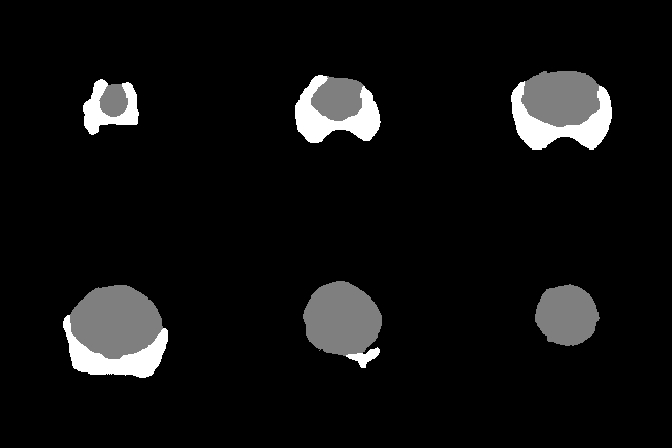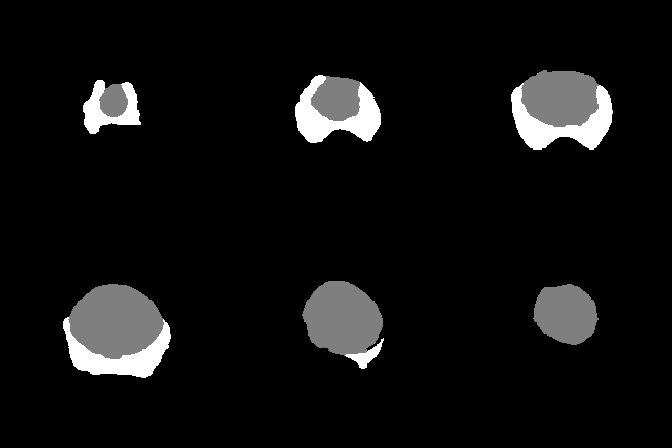A collection of prostate MR studies with expert annotations
Prostate158 is a curated dataset of biparametric 3 Tesla prostate MRI images for automatic segmentation of anatomical zones and cancerous lesions. Histopathologic confirmation is available for each cancerous lesion. All studies include a T2-weighted (T2W) and diffusion-weighted (DWI) images with apparent diffusion coefficient (ADC) maps. Images in each study were resampled so that orientation, direction, and spacing were the same.
The training dataset (139 MRIs) is freely available at .
The test dataset consists of 19 additional MRIs and can be found at
We have trained U-Net base models for segmentation of the anatomical zones as well as the cancer areas in the prostate images, which can serve as a basis for comparison with future approaches. The weights for the models are available at:
Since two raters segmented the anatomical zones and the cancer areas in the test dataset, the model score is given in comparison to each rater.
| Rater 1 | Rater 2 | ||||||
| Metric | Transitional Zone | Peripheral Zone | Cancer | Transitional Zone | Peripheral Zone | Cancer | |
|---|---|---|---|---|---|---|---|
| Dice Coefficient | 0.877 | 0.754 | 0.453 | 0.875 | 0.730 | 0.398 | |
| Hausdorff Distance | 18.3 | 22.8 | 36.7 | 17.5 | 33.2 | 39.5 | |
| Surface Distance | 2.19 | 1.95 | 17.37 | 2.59 | 1.88 | 19.13 | |
The baseline has been trained with the following packages and versions:
MONAI version: 0.9.dev2149
PyTorch version: 1.9.1
PyTorch Ignite version: 0.4.7
Numpy version: 1.21.2
pandas version: 1.3.5
PyYAML version: 5.4.1
Munch version: 2.5.0
You should be able to install all dependencies with: pip install -U "monai[all]" pyyaml munch pandas
Additionally you should install OpenCV: sudo apt update and sudo apt install libopencv-dev python3-opencv
Adapt the data_dir, the train_csv, valid_csv and test_csv file paths in the anatomy.yaml.
Then execute:
python train.py --config anatomy.yaml
This will launch a training session which should give similar results as in the paper. However, even with a random seed, results between runs my vary up to 1%.
Inside a jupyter notebook, training can be started with the following code:
import monai
from prostate.utils import load_config
from prostate.train import SegmentationTrainer
config = load_config('anatomy.yaml')
monai.utils.set_determinism(seed=config.seed)
trainer=SegmentationTrainer(
config=config,
progress_bar=True,
early_stopping = True,
metrics = ["MeanDice", "HausdorffDistance", "SurfaceDistance"],
save_latest_metrics = True,
)
trainer.run()If satisfied, evaluate the model on a new dataset.
from prostate.data import segmentation_dataloaders
test_dl = segmentation_dataloaders(config=config, train=False, valid=False, test=True)
trainer.evaluate(
checkpoint='models/NAME_OF_CHECKPOINT.pt',
dataloader=test_dl
)See also the train.ipynb notebook for a more detailed tutorial.