EvoMol is a flexible and interpretable evolutionary algorithm designed for molecular properties optimization. It can optimize any (customizable) objective function. It can also maximize the diversity of generated molecules.
EvoMol was designed on Ubuntu (18.04+). Some features might be missing on other systems. Especially, the drawing of exploration trees is currently unavailable on Windows.
To install EvoMol on your system, run the following commands in your terminal. The installation depends on Anaconda.
NOTE (2022/02/02)
In latest versions (v1.5.0+), EvoMol depends on the openbabel python library instead of depending on a "hard"
installation of OpenBabel. Thus, the evomolenv conda environment was changed and it is possible that this causes issues
if you are updating a former EvoMol installation. In the latter case, it is preferable to remove the evomolenv conda
environment (conda remove --name evomolenv --all) and to restart the installation from scratch.
$ git clone https://github.com/jules-leguy/EvoMol.git # Clone EvoMol
$ cd EvoMol # Move into EvoMol directory
$ conda env create -f evomol_env.yml # Create conda environment
$ conda activate evomolenv # Activate environment
$ python -m pip install . # Install EvoMolLaunching a QED optimization for 500 steps. Beware, you need to activate the evomolenv conda environment when you use EvoMol.
from evomol import run_model
run_model({
"obj_function": "qed",
"optimization_parameters": {
"max_steps": 500
},
"io_parameters": {
"model_path": "examples/1_qed"
},
})A dictionary can be given to evomol.run_model to describe the experiment to be performed. This dictionary can contain up to 4 entries, that are described in this section.
Default values are represented in bold.
The "obj_function" attribute can take the following values. Multi-objective functions can be nested to any depth.
- Implemented functions:
- "qed", "plogp", "norm_plogp", "sascore", "norm_sascore", "clscore".
- "isomer_formula" (e.g. "isomer_C7H16").
- "rediscovery_smiles" (e.g. "rediscovery_CC(=O) OC1=CC=CC=C1C(=O)O")
- "homo", "lumo", "gap", "homo-1"
- "entropy_ifg", "entropy_gen_scaffolds", "entropy_shg_1", "entropy_checkmol" and "entropy_ecfp4" can be used to maximize the entropy of descriptors, respectively using IFGs , Murcko generic scaffolds, level 1 shingles, checkmol and ECFP4 fingerprints (RDKit Morgan fingerprints implementation).
- "n_perturbations": count of the number of perturbations that were previously applied on the molecular graph during the optimization. If the "mutation_max_depth" parameter is set to 1, then this is equivalent to the number of mutations.
- "sillywalks_proportion": proportion of ECFP4 features that failed the
sillywalks filter, based on the reference dataset given in the
"silly_molecules_reference_db_path"parameter.
- A custom function evaluating a SMILES. It is also possible to give a tuple (function, string function name).
- A dictionary describing a multi-objective function and containing the following entries (see the example section).
"type":- "linear_combination" (linear combination of the properties).
- "product" (product of the properties).
- "mean" (mean of the properties).
- "abs_difference" (absolute difference of exactly 2 properties).
"functions": list of functions (string keys describing implemented functions, custom functions, multi-objective functions or wrapper functions).- Specific to the linear combination
"coef": list of coefficients.
- A dictionary describing a function wrapping a single property and containing the following entries (see the example section).
"type":- "gaussian" (passing the value of a unique objective function through a Gaussian function).
- "opposite" (computing the opposite value of a unique objective function).
- "sigm_lin", (passing the value of a unique objective through a linear function and a sigmoid function).
- "one_minus" (computing 1-f(x) of a unique objective function f).
"function"the function to be wrapped (string key describing an implemented function, custom function, multi_objective function or wrapper function). For compatibility reasons, it is also possible to use a"functions"attribute that contains a list of functions. In that case only the first element of the list is considered.- Specific to the use of a Gaussian function
"mu": μ parameter of the Gaussian."sigma": σ parameter of the Gaussian."normalize": whether to normalize the function so that the maximum value is exactly 1 (False).
- Specific to the use of sigmoid/linear functions
"a"list of a coefficients for the ax+b linear function definition."b"list of b coefficients for the ax+b linear function definition."lambda"list of λ coefficients for the sigmoid function definition.
- An instance of evomol.evaluation.EvaluationStrategyComposant
"guacamol_v2"for taking the goal directed GuacaMol benchmarks.
The "action_space_parameters" attribute can be set with a dictionary containing the following entries.
"atoms": text list of available heavy atoms ("C,N,O,F,P,S,Cl,Br")."max_heavy_atoms": maximum molecular size in terms of number of heavy atoms (38)."append_atom": whether to use append atom action (True)."remove_atom": whether to use remove atom action (True)."change_bond": whether to use change bond action (True)."change_bond_prevent_breaking_creating_bonds": whether to prevent the removal or creation of bonds by change_bond action (False)"substitution": whether to use substitute atom type action (True)."cut_insert": whether to use cut atom and insert carbon atom actions (True)."move_group": whether to use move group action (True)."remove_group": whether to use remove group action (False)."remove_group_only_remove_smallest_group": in case remove group action is enabled, whether to be able to remove- both parts of a bridge bond (False), or only the smallest part in number of atoms (True).
"use_rd_filters": whether to use the rd_filter program as a quality filter before inserting the mutated individuals in the population (False)."sillywalks_thresholdmaximum proportion of silly bits in the ECFP4 fingerprint of the solutions with respect to a reference dataset (see IO parameters). If the proportion is above the threshold, the solutions will be discarded and thus will not be inserted in the population (1)."sascore_threshold"if the solutions have a SAScore value above this threshold, they will be discarded and thus will not be inserted in the population (float("inf"))."custom_filter_function": custom boolean function evaluating a SMILES that defines a filter on the accessible search space. If the result isTrue, the molecule is considered valid, and otherwise it is considered invalid (None)."sulfur_valence": valence of sulfur atoms (6).
The "optimization_parameters" attribute can be set with a dictionary containing the following entries.
"pop_max_size": maximum population size (1000)."max_steps": number of steps to be run before stopping EvoMol (1500)."max_obj_calls"": number of calls to the objective functions before stopping EvoMol (float("inf"))."stop_kth_score_value": stopping the search if the kth score in descendant value order has reached the given value with given precision. Accepts a tuple (k, score, precision) or None to disable."k_to_replace": number of individuals replaced at each step (10)."selection": whether the best individuals are selected to be mutated ("best"), or they are selected randomly with uniform distribution ("random"), or they are selected randomly with a probability that is proportional to their objective function value ("random_weighted") ."problem_type": whether it is a maximization ("max") or minimization ("min") problem."mutation_max_depth": maximum number of successive actions on the molecular graph during a single mutation (2)."neighbour_generation_strategy": strategy to generate neighbour candidates (mutation). By default, the type of perturbation is first drawn randomly, then the actual perturbation of previously selected type is drawn randomly among the valid ones (evomol.molgraphops.exploration.RandomActionTypeSelectionStrategy())."mutation_find_improver_tries": maximum number of mutations to find an improver (50)."guacamol_init_top_100": whether to initialize the population with the 100 best scoring individuals of the GuacaMol ChEMBL subset in case of taking the GuacaMol benchmarks (False). The list of SMILES must be given as initial population."mutable_init_pop": if True, the individuals of the initial population can be freely mutated. If False, they can be branched but their atoms and bonds cannot be modified (True)."n_max_desc": max number of descriptors to be possibly handled when using an evaluator relying on a vector of descriptors such as entropy contribution (3.000.000)."shuffle_init_pop": whether to shuffle the smiles at initialization (False).
The "io_parameters" attribute can be set with a dictionary containing the following entries.
"model_path": path where to save model's output data ("EvoMol_model")."smiles_list_init": list of SMILES describing the initial population (None: interpreting the"smiles_list_init_path"attribute). Note : not available when taking GuacaMol benchmarks."smiles_list_init_path": path where to find the SMILES list text file describing the initial population (one SMILES per row). It is also possible to pass the path to the pop.csv file from a previous EvoMol experiment. In the latter case, the population will be initialized as it was at the end of the loaded experiment (None: initialization of the population with a single methane molecule)."external_tabu_list": list of SMILES that won't be generated by EvoMol."record_history": whether to save exploration tree data. Must be set to True to later draw the exploration tree (False)."record_all_generated_individuals": whether to record a list of all individuals that are generated during the entire execution (even if they fail the objective function computation or if they are not inserted in the population as they are not improvers). Also recording the step number and the total number of calls to the objective function at the time of generation (False)."save_n_steps": period (steps) of saving the data (100)."print_n_steps": period (steps) of printing current population statistics (1)."dft_working_dir": path where to save DFT optimization related files ("/tmp")."dft_cache_files": list of json files containing a cache of previously computed HOMO or LUMO values ([])."dft_MM_program": program used to compute molecular mechanics initial geometry of DFT calculations. The options are :- "obabel_mmff94" or "obabel" to combine OpenBabel and the MMFF94 force field.
- "rdkit_mmff94" to combine RDKit with the MMFF94 force field.
- "rdkit_uff" to combine RDKit with the UFF force field.
"dft_base": DFT calculations base ("3-21G*")."dft_method": DFT calculations method (B3LYP)."dft_n_jobs": number of threads assigned to each DFT calculation (1)."dft_mem_mb": memory assigned to each DFT calculation in MB (512)."silly_molecules_reference_db_path: path to a JSON file that represents a dictionary containing as keys all the ECFP4 bits that are extracted from a reference dataset of quality solutions (None). See the"sillywalks_threshold"parameter."evaluation_strategy_parameters": a dictionary that contains an entry "evaluate_init_pop" to set given parameters to the EvaluationStrategy instance in the context of the evaluation of the initial population. An entry "evaluate_new_sol" must be also contained to set given parameters for the evaluation of new solutions during the optimization process. If None, both keys are set to an empty set of parameters (None).
In order to aggregate several objective functions into a single function that will be optimized by EvoMol, it is
neccessary to use the dictionary (tree) function declaration for the "obj_function" attribute.
As an example, we design here an objective function to solve a virtual problem. The problem is to find a molecule with a QED value of about 0.8, a CLScore value above 4, and about 70% of carbon atoms (ignoring the hydrogen atoms).
First, we design a function that counts the proportion of heteroatoms.
from rdkit.Chem import Lipinski, MolFromSmiles
def hetero_atoms_proportion(smiles):
return Lipinski.NumHeteroatoms(MolFromSmiles(smiles)) / Lipinski.HeavyAtomCount(MolFromSmiles(smiles))Then, the objective function can be designed this way. We consider that the most important property here is the QED value (coefficient 0.5) and that the two other properties are equally important (coefficient 0.25). The sum of the coefficients may be different from 1, but since all the functions range between 0 and 1 here, this allows the final function to also range between 0 and 1.
objective_tree = {
# The objective function is a linear combination of three sub-functions
"type": "linear_combination",
"coef": [0.5, 0.25, 0.25],
"functions": [
{
# Centering a Gaussian function on the 0.8 QED value
"type": "gaussian",
"function": "qed",
"mu": 0.8,
"sigma": 0.5,
"normalize": True
},
{
# The parameters of the sigmoid function are chosen so that the function value is above 0.99 when the CLScore value is above 4
"type": "sigm_lin",
"function": "clscore",
"a": -1,
"b": 3.5,
"lambda": 10
},
{
# Centering a Gaussian function on the 0.7 proportion of hetero atoms
"type": "gaussian",
"function": {
# Returning 1 - (proportion of heteroatoms) to represent the proportion of carbon atoms
"type": "one_minus",
"function": (hetero_atoms_proportion, "hetero_atoms_proportion")
},
"mu": 0.7,
"sigma": 0.5,
"normalize": True
},
]
}The designed objective function can then be optimized by calling the run_model function of EvoMol
from evomol import run_model
run_model({
"obj_function": objective_tree,
"io_parameters": {
"model_path": "examples/multiobjective_run"
}
})Performing a QED optimization run of 500 steps, while recording the exploration data.
from evomol import run_model
model_path = "examples/2_large_tree"
run_model({
"obj_function": "qed",
"optimization_parameters": {
"max_steps": 500},
"io_parameters": {
"model_path": model_path,
"record_history": True
}
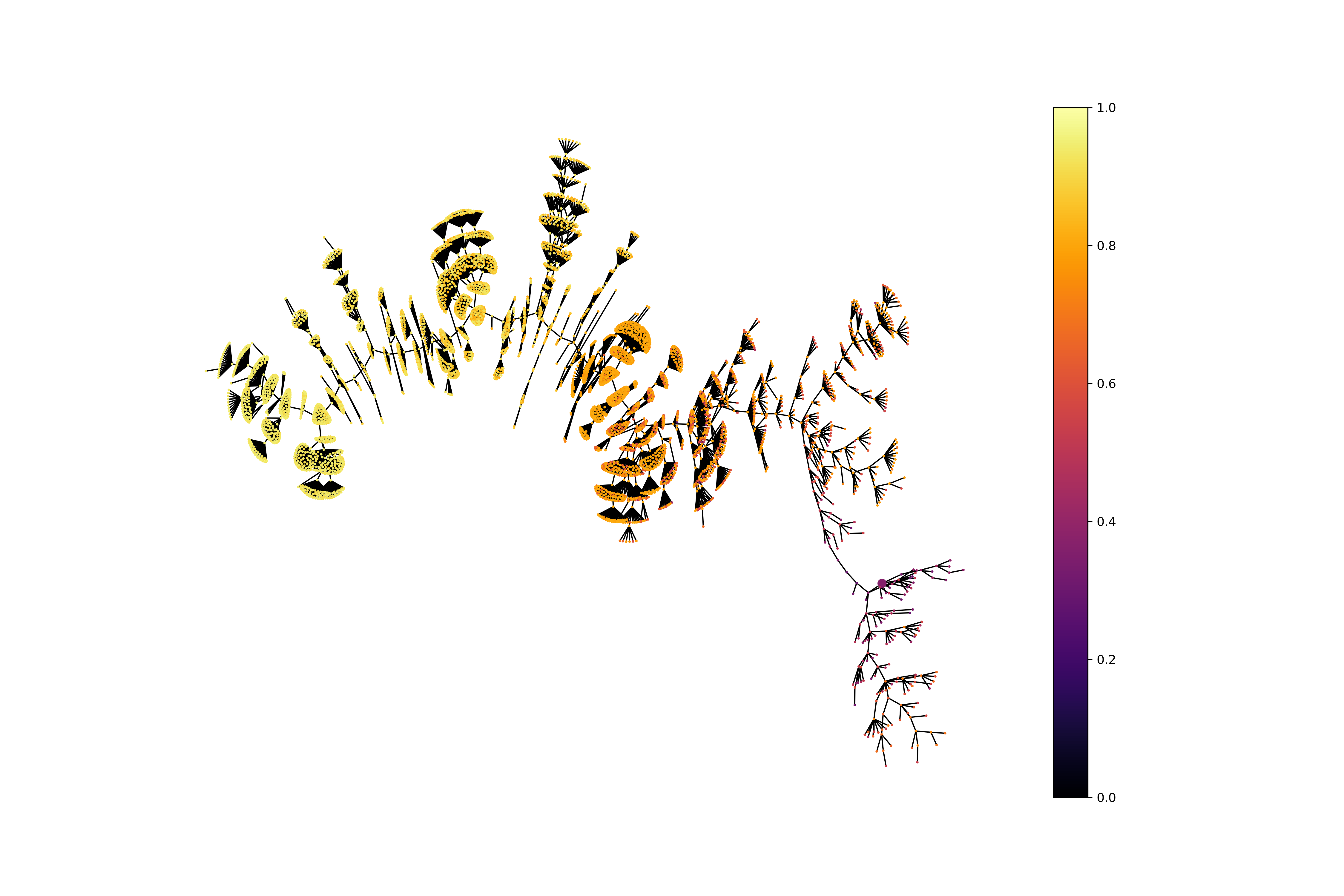
})Plotting the exploration tree with solutions colored according to their score. Nodes represent solutions. Edges represent mutations that lead to an improvement in the population.
from evomol.plot_exploration import exploration_graph
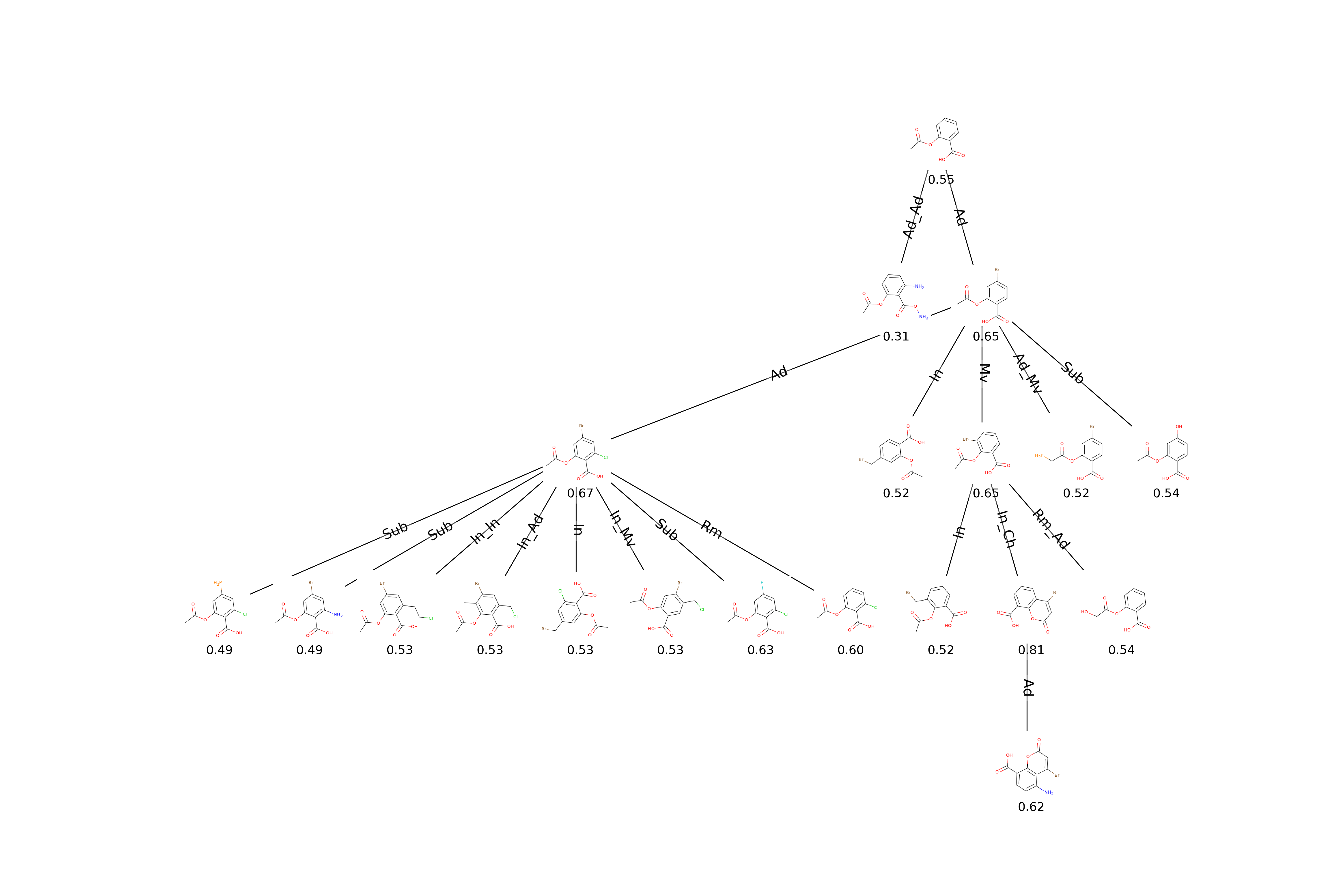
exploration_graph(model_path=model_path, layout="neato")Performing the experiment of mutating a fixed core of acetylsalicylic acid to increase its QED value.
from evomol import run_model
model_path = "examples/3_detailed_tree"
run_model({
"obj_function": "qed",
"optimization_parameters": {
"max_steps": 10,
"pop_max_size": 10,
"k_to_replace": 2,
"mutable_init_pop": False
},
"io_parameters": {
"model_path": model_path,
"record_history": True,
"smiles_list_init_path": "examples/acetylsalicylic_acid.smi"
}
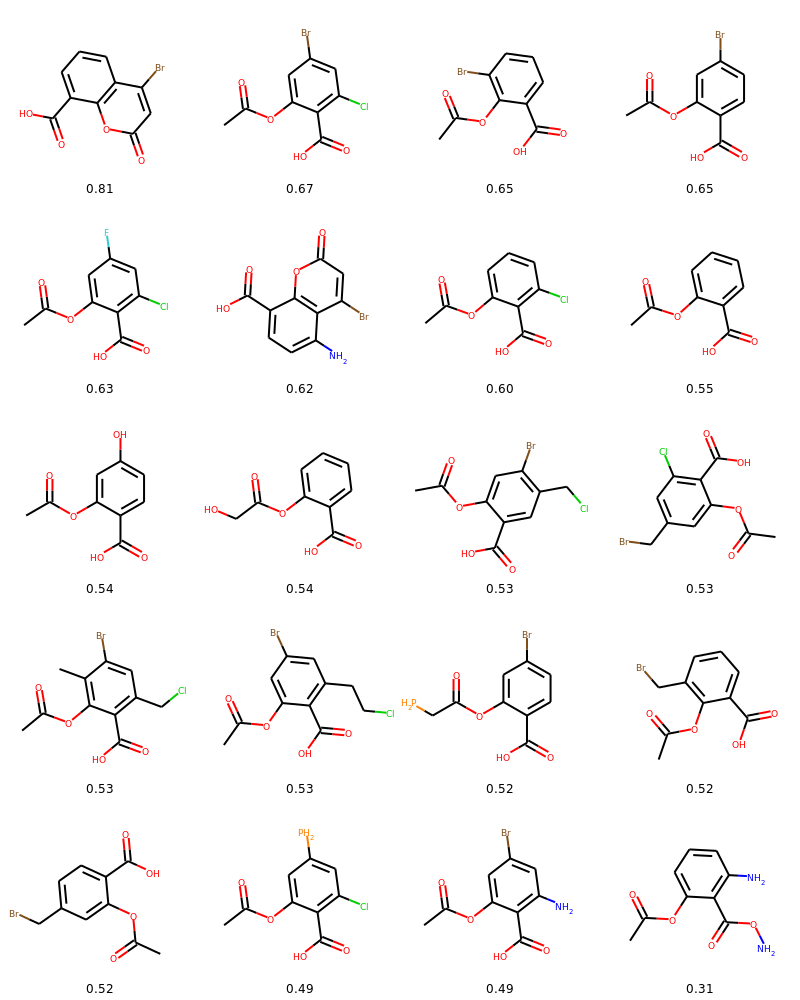
})Plotting the exploration tree including molecular drawings, scores and action types performed during mutations. Also plotting a table of molecular drawings.
from evomol.plot_exploration import exploration_graph
exploration_graph(model_path=model_path, layout="dot", draw_actions=True, plot_images=True, draw_scores=True,
root_node="O=C(C)Oc1ccccc1C(=O)O", legend_scores_keys_strat=["total"], mol_size_inches=0.3,
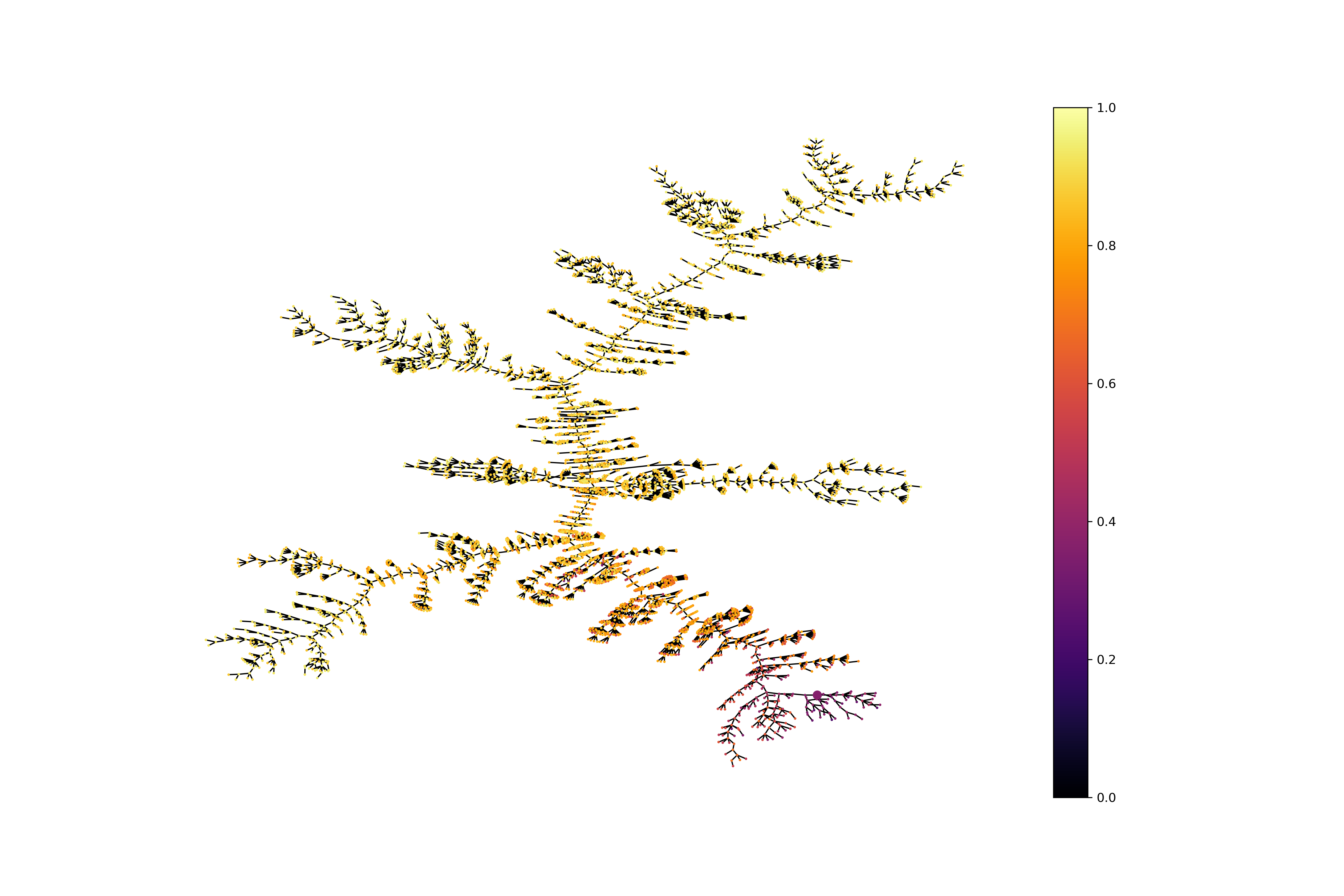
mol_size_px=(800, 800), legend_offset=(-0.007, -0.05), figsize=(20, 20/1.5), legends_font_size=13)Optimizing jointly the QED and the entropy of IFGs using a linear combination. The weights are set respectively to 1 and 10.
from evomol import run_model
model_path = "examples/4_entropy_optimization"
run_model({
"obj_function": {
"type": "linear_combination",
"functions": ["qed", "entropy_ifg"],
"coef": [1, 10]
},
"optimization_parameters": {
"max_steps": 500,
"pop_max_size": 1000
},
"io_parameters": {
"model_path": model_path,
"record_history": True
},
})Plotting the exploration trees representing the QED values.
from evomol.plot_exploration import exploration_graph
exploration_graph(model_path=model_path, layout="neato", prop_to_study_key="qed")It is possible to use the dictionary based declaration of the objective function to evaluate molecules independently of
the optimization procedure defined by EvoMol. The evomol.get_objective_function_instance function is very
comparable to evomol.run_model, but returns an object that can be used to evaluate any molecule.
The returned object is an instance of evomol.evaluation.EvaluationStrategyComposant, which has in particular an
eval_smi method that can evaluate any molecule represented as a SMILES string.
The input of evomol.get_objective_function_instance is a dictionary that can contain the following attributes
"obj_function": definition of the evaluation function (see relevant section)."io_parameters": dictionary that is used here to define the parameters relative to DFT and molecular mechanics parameters (see relevant section)."optimization_parameters": dictionary that is used here to define the parameters relative to entropy contribution evaluation ("n_max_desc"and"pop_max_size"parameters defined here).
For a more complete use of the evaluation function, see this tutorial .
As the CLscore is dependent of prior
data to be computed, EvoMol needs to be given the data location. To do so, the $SHINGLE_LIBS environment variable
must be set to the location of the shingle_libs folder that can be
downloaded here.
To perform DFT computations, you need to bind Gaussian09 or another version of Gaussian with EvoMol.
To do so, the $OPT_LIBS environment variable must point to a folder containing run.sh, a script launching a
DFT optimization with Gaussian09 of the input filepath given as parameter.
Here is an example of such a file.
#!/usr/bin/env bash
export g09root=/path/to/the/folder/that/contains/g09/installation/folder
source g09root/g09/bsd/g09.profile
g09 $1 $2In order to use the checkmol descriptor for entropy evaluation, the $CHECKMOL_EXE environment variable must point
to the executable of the
checkmol program.
To use EvoMol for GuacaMol goal directed benchmarks optimization using the best scoring molecules from their subset of ChEMBL as initial population, you need to :
- Download the ChEMBL subset.
- Give the path of the data using the
"smiles_list_init_path"attribute. - Insure that the
"guacamol_init_top_100"attribute is set to True.
To use the rd_filter program as a filter of solutions that can be
inserted in the population, the $FILTER_RULES_DATA environment variable must point to a folder containing the
rules.json and alert_collection.csv files.
In order to use advanced functions of EvoMol, some tutorials are available in the
tutorials folder.
The tutorials currently available are :
- Advanced use of the evaluation functions outside EvoMol
.
- Declaration of evaluation functions that can be used to evaluate molecules outside EvoMol, based on the tree-based dictionary declaration for the objective functions of EvoMol.
- Description of the evomol.evaluation.EvaluationStrategyComposant object and its methods that allow the user to obtain the scores and sub-scores values for arbitrary molecules.
- Starting a new EvoMol optimization where a previous one stopped
- Starting an EvoMol optimization procedure using as initial population the final population of a previous experiment.
- Building a cache of DFT-dependent properties values in the previous experiment so that they are not evaluated again in the initial population of the new experiment.
To reference EvoMol, please cite one of the following articles.
Leguy, J., Cauchy, T., Glavatskikh, M., Duval, B., Da Mota, B. EvoMol: a flexible and interpretable evolutionary algorithm for unbiased de novo molecular generation. J Cheminform 12, 55 (2020). https://doi.org/10.1186/s13321-020-00458-z
Leguy, J., Glavatskikh, M., Cauchy, T. et al. Scalable estimator of the diversity for de novo molecular generation resulting in a more robust QM dataset (OD9) and a more efficient molecular optimization. J Cheminform 13, 76 (2021). https://doi.org/10.1186/s13321-021-00554-8