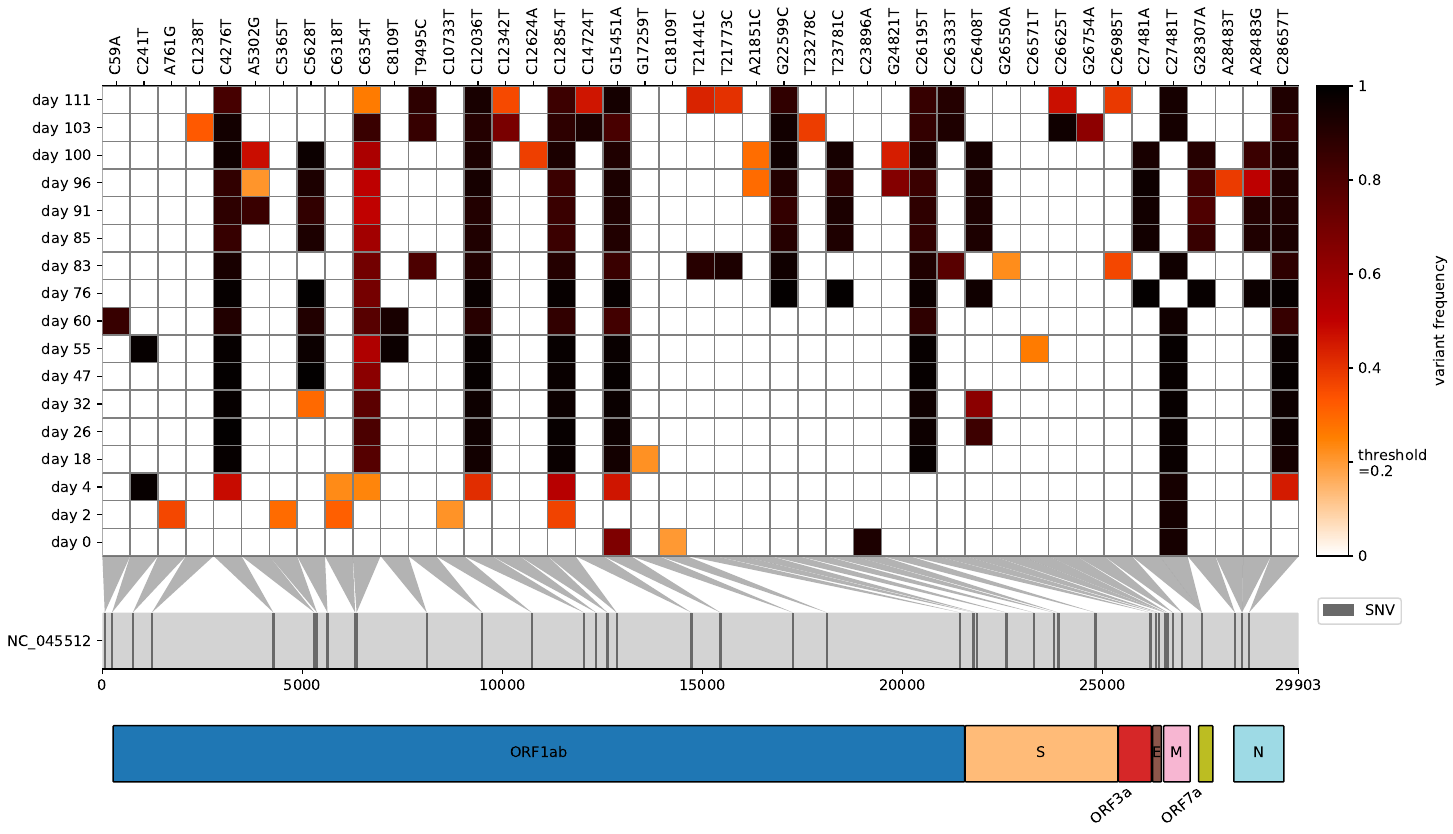
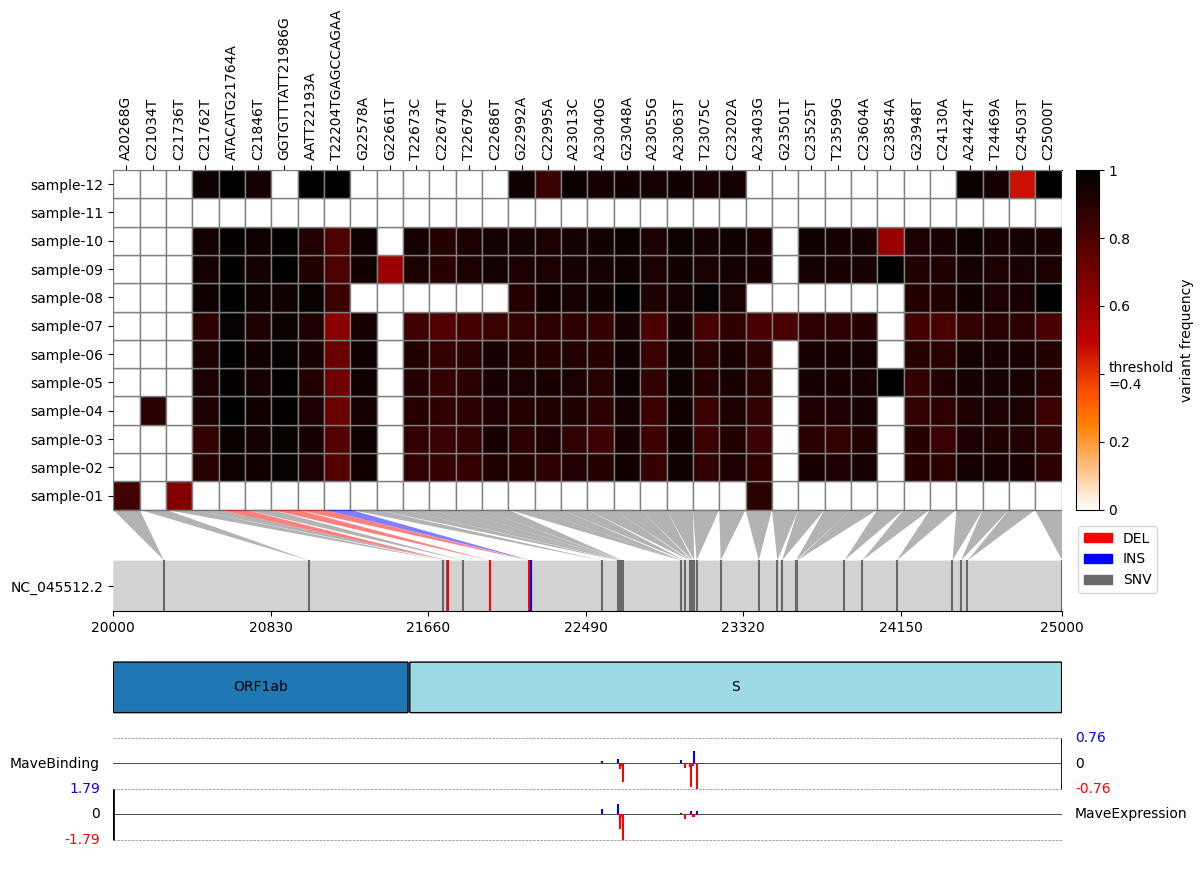
virHEAT is a tool to visualize vcfs as a heatmap and map mutations to respective genes.
Ever wanted to have a condensed look at variant frequencies after mapping your raw reads to a viral/bacterial reference genome and compare multiple vcf files at the same time? Than virHEAT is for you. You can not only visualize the heatmap but also read in a gff3 file that lets you display genes harboring a mutation. This lightweight script was inspired by snipit and my variant frequency plot, getting the best visualization features of both.
pip install virheatconda install -c bioconda virheatgit clone https://github.com/jonas-fuchs/virHEAT
cd virHEAT
pip install -r requirements.txt
# or
pip install .That was already it. To check if it worked:
virheat -vYou should see the current virHEAT version.
usage: virheat <folder containing vcfs> <output dir> -l or -g [additional arguments]
Arguments:
positional arguments:
input folder containing input files and output folder
options:
-h, --help show this help message and exit
-r ref_id, --reference ref_id
reference identifier
--name virHEAT_plot.pdf
plot name and file type (pdf, png, svg, jpg). Default: virHEAT_plot.pdf
-l None, --genome-length None
length of the genome (needed if gff3 is not provided)
-g None, --gff3-path None
path to gff3 (needed if length is not provided)
-a [gene ...], --gff3-annotations [gene ...]
annotations to display from gff3 file (standard: gene). Multiple possible.
-t 0, --threshold 0 display frequencies above this threshold (0-1)
--delete, --no-delete
delete mutations that are present in all samples and their maximum frequency divergence is smaller than 0.5 (default: True)
-n None, --delete-n None
do not show mutations that occur n times or less (default: Do not delete)
-z start stop, --zoom start stop
restrict the plot to a specific genomic region.
--sort, --no-sort sort sample names alphanumerically (default: False)
--min-cov 20 display mutations covered at least x time (only if per base cov tsv files are provided)
-s scores_file pos_col score_col score_name, --scores scores_file pos_col score_col score_name
specify scores to be added to the plot by providing a CSV file containing scores, along with its column for amino-acid positions, its column for scores, and descriptive score names (e.g., expression, binding, antibody escape, etc.).
This option can be used multiple times to include multiple sets of scores.
-v, --version show program's version number and exit
You need to either provide the length of your reference genome or if you want to get the sequence annotation you will need to provide the gff3 file.
Additionally, you can also analyse if mutations are sufficiently covered and display non-covered cells in grey. For that first create a per base coverage tsv files for each bam file with Qualimap and provide it in the same folder as the vcf files. Give them the same name as your vcf files.
Moreover, there is an option to include visualizations of additional scores (e.g., MAVE scores for binding affinity, expression level, antibody escape, etc.) mapped to mutations on the heatmap. To utilize this feature, use the -s or --scores argument, and provide the following arguments: 1) path to the CSV file containing scores; 2) the name of the column in this file containing mutation positions in classic notation (e.g., T430Y); 3) the name of the column in this file containing the scores themselves; 4) a descriptive score name that will be used as labels in the plot. Multiple score sets can be included simultaneously by repeating the -s or --scores option with different arguments. For example input and possible output data, please refer to the files located in the example_data/example_mave_data folder.
Important disclaimer: The code is under the GPLv3 licence. The code is WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.