The early detection of skin cancer substantially improves the five-year survival rate of patients. It is often difficult to distinguish early malignant tumors from skin images, even by expert dermatologists. Therefore, several classification methods of dermatoscopic images have been proposed, but they have been found to be inadequate or defective for skin cancer detection, and often require a large amount of calculations. This study proposes an improved capsule network called FixCaps for dermoscopic image classification. FixCaps has a larger receptive field than CapsNets by applying a high-performance large-kernel at the bottom convolution layer whose kernel size is as large as 31
https://doi.org/10.1109/ACCESS.2022.3181225
Note: The augmented data of HAM10000 can be obtained as follows: https://aistudio.baidu.com/aistudio/datasetdetail/151696
- Classification accuracy (%) on the HAM10000 test set.
| Method | Accuracy [%] | Params(M) | FLOPs(G) |
|---|---|---|---|
| GoogLeNet | 83.94 | 5.98 | 1.58 |
| Inception V3 | 86.82 | 22.8 | 5.73 |
| MobileNet V3 | 89.97 | 1.53 | 0.12 |
| IRv2-SA | 93.47 | 47.5 | 25.46 |
| FixCaps-DS | 96.13 | 0.14 | 0.08 |
| FixCaps | 96.49 | 0.5 | 6.74 |
- The accuracy is evaluated on the test set by using different LKC(large-kernel convolution).
3 Evaluation metrics of the FixCaps.
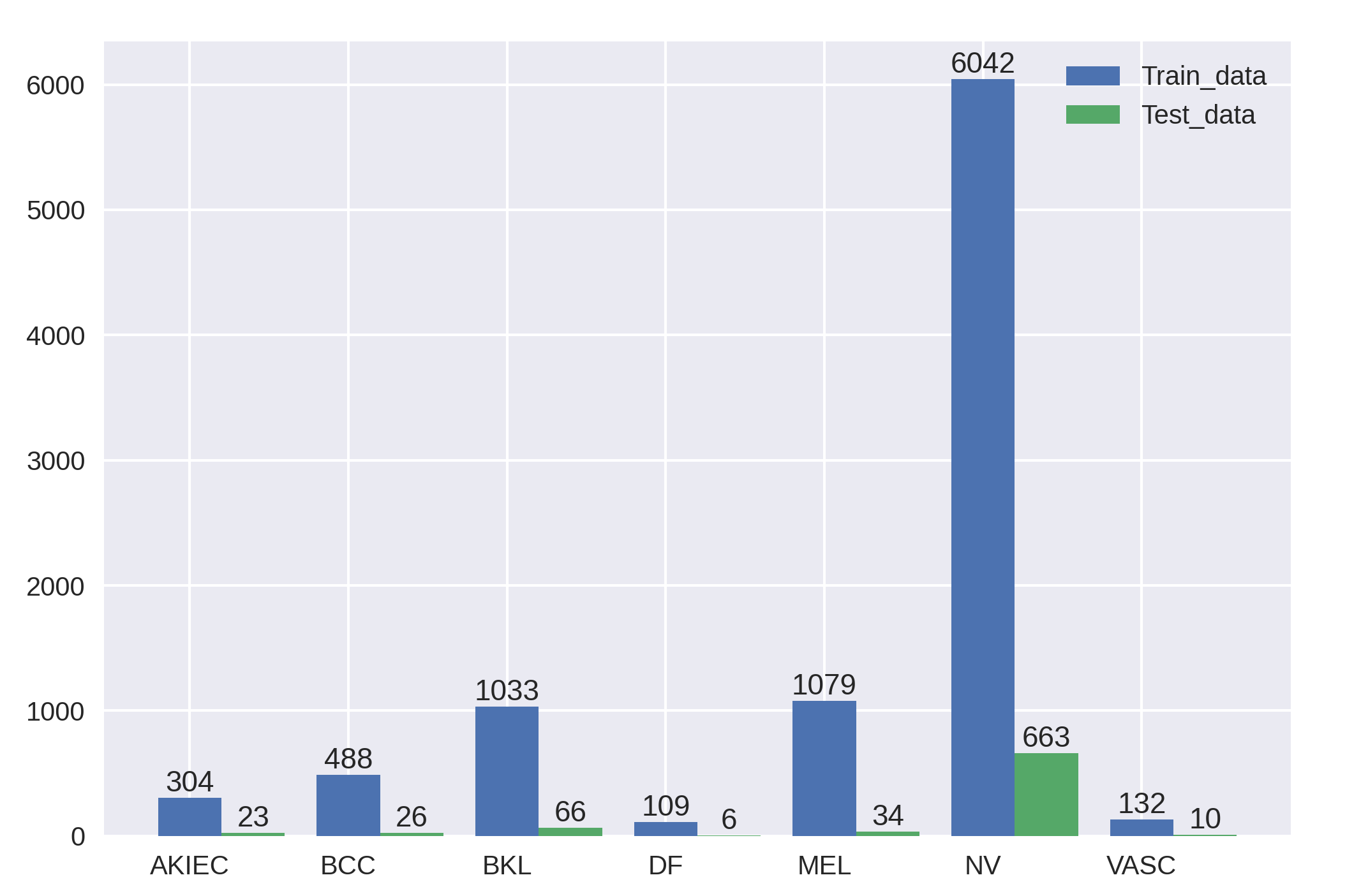
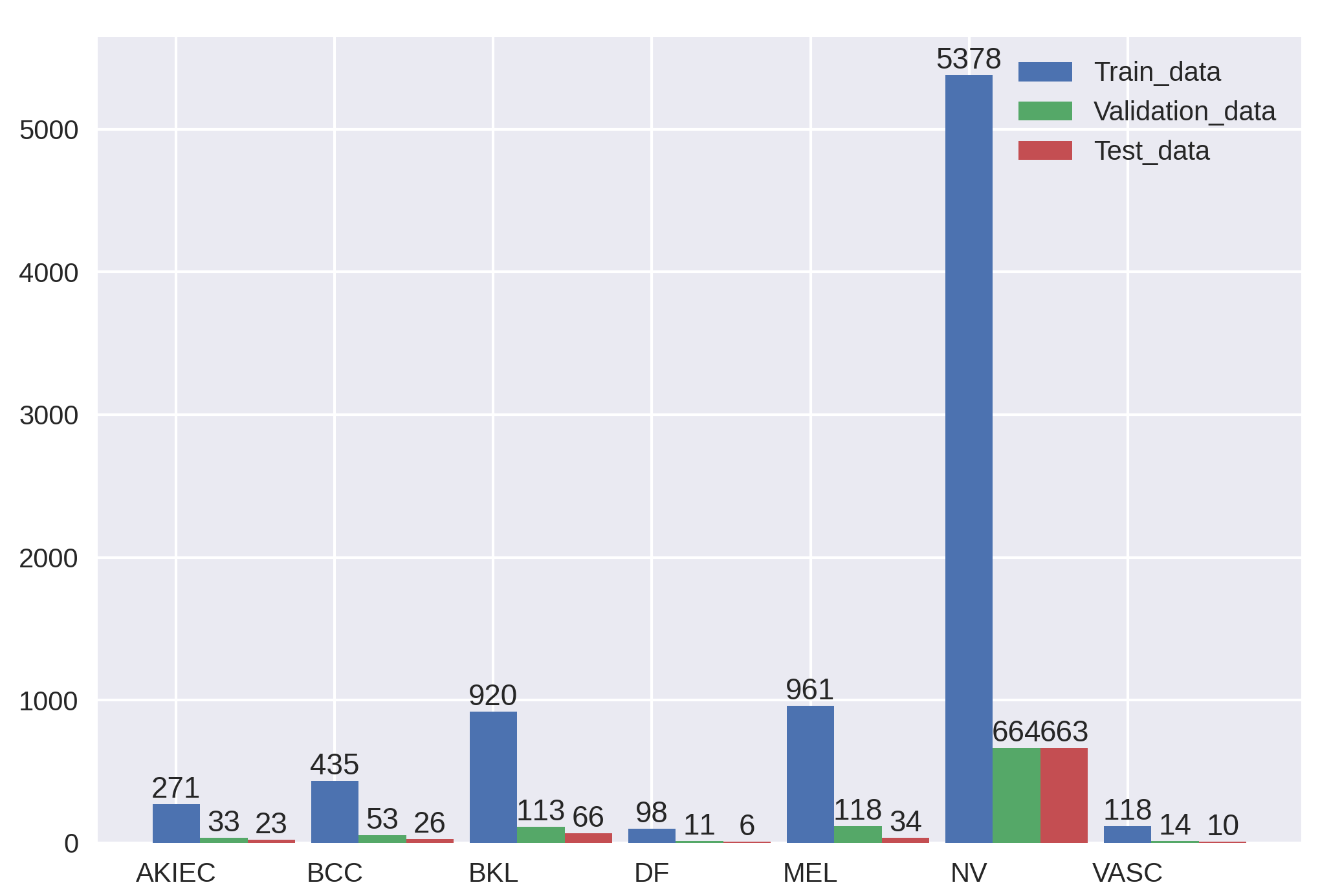
| FixCaps-31 | Distribution of the HAM10000 Dataset | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
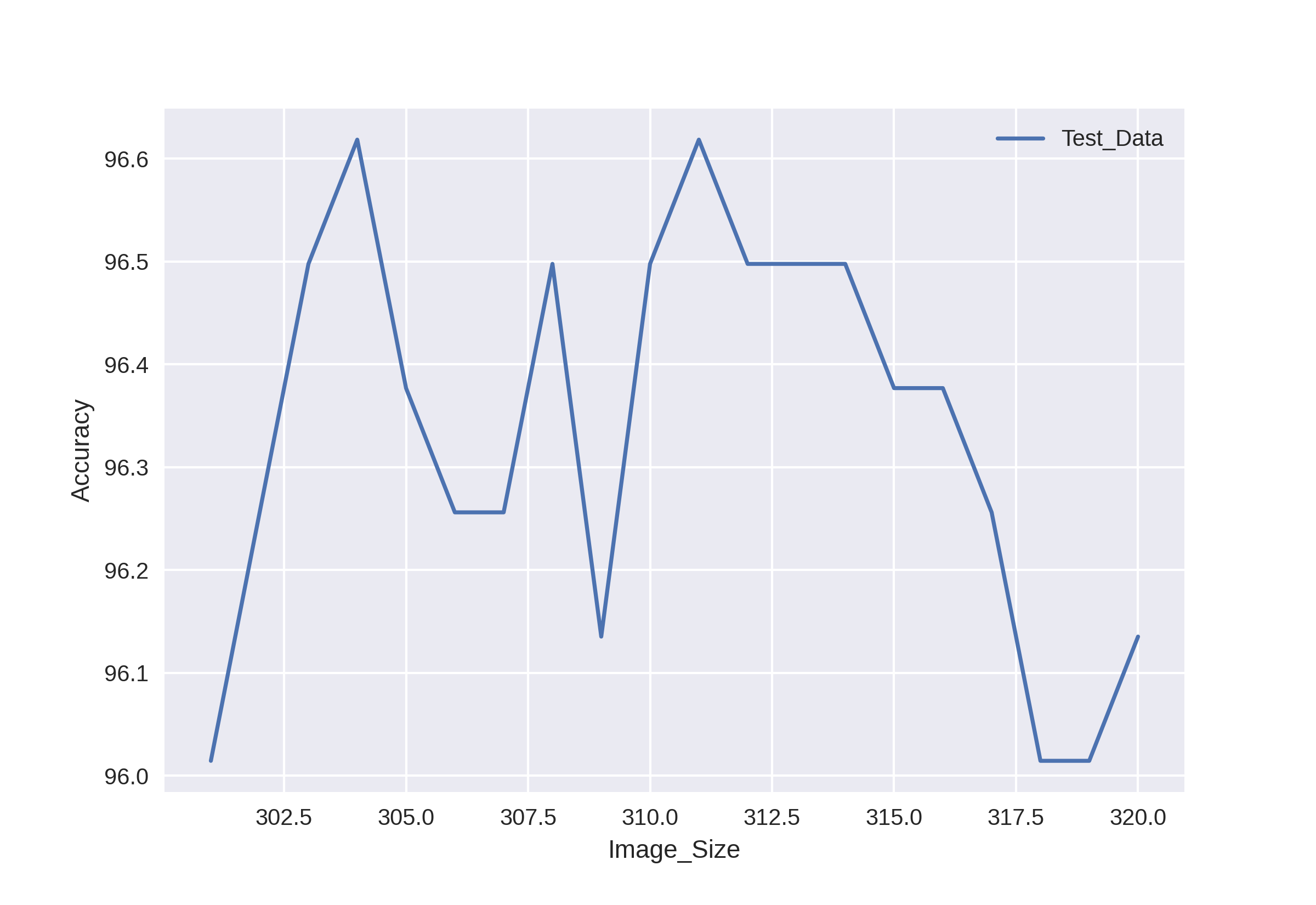
4 Generalization Performance
| Robustness(FixCaps-29) | Distribution of the HAM10000 Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FixCaps-29 | Evaluation Metrics(RTX3070_Driver_Version: 515.7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FixCaps-18(Ablation-CAM) | FixCaps_DS-18 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
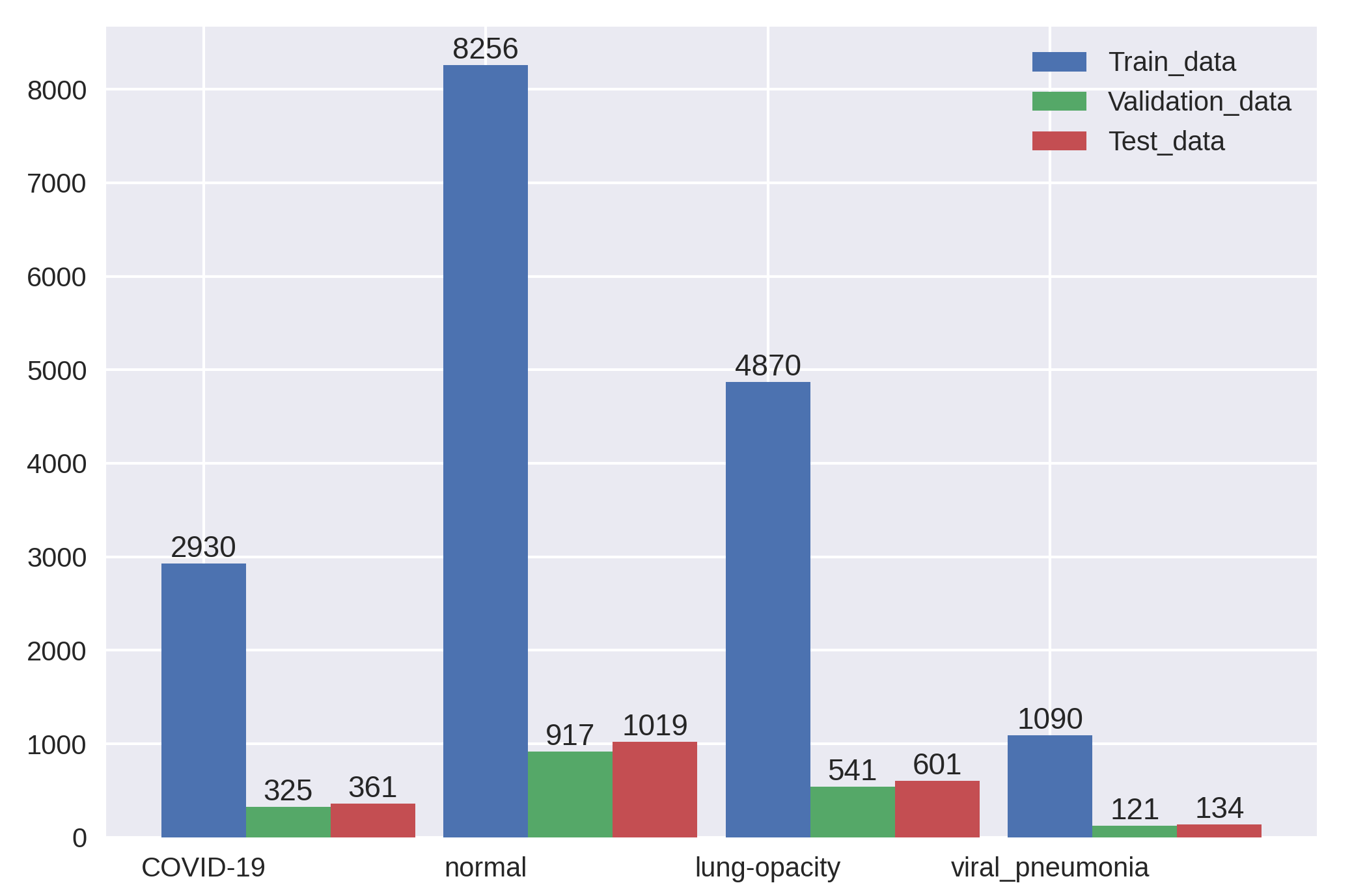
Dataset: https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database
The COVID-19 Radiography Database consisted of 21165 images.
Among them, covid(3616),normal(10192),opacity(6012),viral(1345).
| Evaluation Metrics | Distribution of the COVID-19 Radiography Dataset | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Source Data: https://dx.doi.org/10.5281/zenodo.1214456
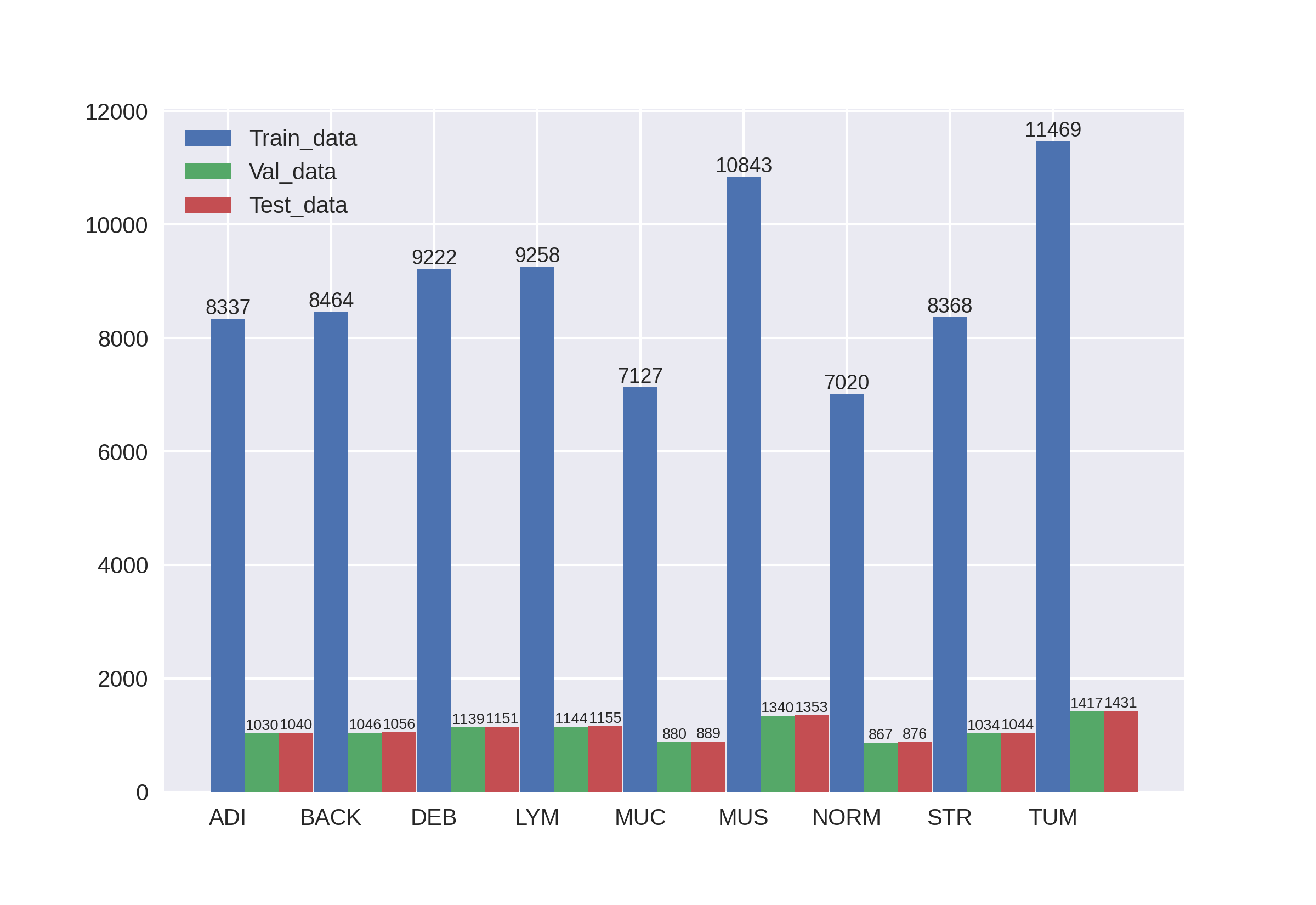
Jakob Nikolas Kather, Johannes Krisam, et al., "Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study," PLOS Medicine, vol. 16, no. 1, pp. 1–22, 01 2019.
This is a slightly different version of the "NCT-CRC-HE-100K" image set: This set contains 100,000 images in 9 tissue classes at 0.5 MPP and was created from the same raw data as "NCT-CRC-HE-100K".
However, no color normalization was applied to these images. Consequently, staining intensity and color slightly varies between the images. Please note that although this image set was created from the same data as "NCT-CRC-HE-100K", the image regions are not completely identical because the selection of non-overlapping tiles from raw images was a stochastic process.
| FixCaps-DS-18 | NCT-CRC-HE-100K-NONORM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Example of Skin lesions in HAM10000 dataset.
Among them, BKL, DF, NV, and VASC are benign tumors, whereas AKIEC, BCC, and MEL are malignant tumors.
Available:
https://challenge.isic-archive.com/data/#2018
https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/DBW86T
HAM10000 dataset:
Noel Codella, Veronica Rotemberg, Philipp Tschandl, M. Emre Celebi, Stephen Dusza, David Gutman, Brian Helba, Aadi Kalloo, Konstantinos Liopyris, Michael Marchetti, Harald Kittler, Allan Halpern: "Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC)", 2018; https://arxiv.org/abs/1902.03368
Tschandl, P., Rosendahl, C. & Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 5, 180161 doi:10.1038/sdata.2018.161 (2018).
Available: https://www.nature.com/articles/sdata2018161, https://arxiv.org/abs/1803.10417
The dataset is released under a Creative Commons Attribution 4.0 License. For more information, see https://creativecommons.org/licenses/by/4.0/ .
a. IRv2-SA
S. K. Datta, M. A. Shaikh, S. N. Srihari, and M. Gao. "Soft-Attention Improves Skin Cancer Classification Performance,"
Computer Science, vol 12929. Springer, Cham, 2021. doi: 10.1007/978-3-030-87444-5_2.
https://github.com/skrantidatta/Attention-based-Skin-Cancer-Classification
b. SLA-StyleGAN
C. Zhao, R. Shuai, L. Ma, W. Liu, D. Hu and M. Wu, ``Dermoscopy Image Classification Based on StyleGAN and DenseNet201,"
in IEEE Access, vol. 9, pp. 8659-8679, 2021, doi: 10.1109/ACCESS.2021.3049600.
If you use FixCaps for your research or aplication, please consider citation:
@ARTICLE{9791221,
author={Lan, Zhangli and Cai, Songbai and He, Xu and Wen, Xinpeng},
journal={IEEE Access},
title={FixCaps: An Improved Capsules Network for Diagnosis of Skin Cancer},
year={2022},
volume={10},
number={},
pages={76261-76267},
doi={10.1109/ACCESS.2022.3181225}
}
@ARTICLE{LanCapsNets,
author={Lan, Zhangli and Cai, Songbai and Zhu, Jiqiang and Xu, Yuantong},
journal={XXX on XXX},
title={A Novel Skin Cancer Assisted Diagnosis Method based on Capsule Networks with CBAM},
year={},
volume={},
number={},
pages={},
doi={10.36227/techrxiv.23291003},
}