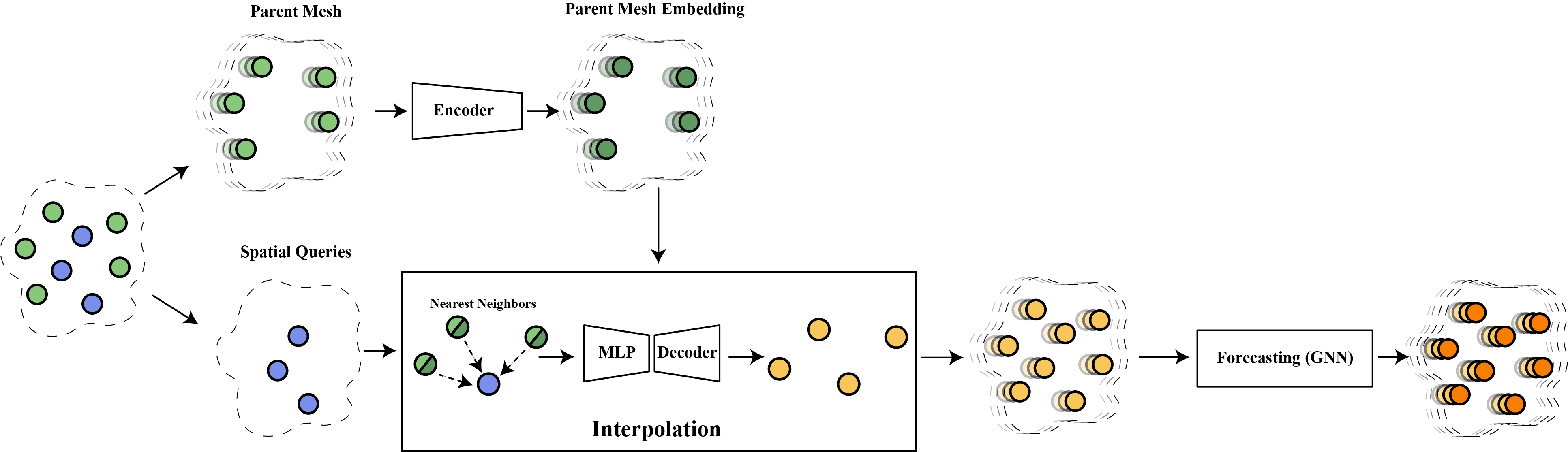
This is the official repository to the paper "MAgNet: Mesh-Agnostic Neural PDE Solver" by Oussama Boussif, Dan Assouline, and professors Loubna Benabbou and Yoshua Bengio.
In this paper, we aim to address the problem of learning solutions to Partial Differential Equations (PDE) while also generalizing to any mesh or resolution at test-time. This effectively enables us to generate predictions at any point of the PDE domain.
To cite our work, please use the following bibtex:
@inproceedings{magnet_neurips_2022,
author = {Boussif, Oussama and Bengio, Yoshua and Benabbou, Loubna and Assouline, Dan},
booktitle = {Advances in Neural Information Processing Systems},
editor = {S. Koyejo and S. Mohamed and A. Agarwal and D. Belgrave and K. Cho and A. Oh},
pages = {31972--31985},
publisher = {Curran Associates, Inc.},
title = {MAgNet: Mesh Agnostic Neural PDE Solver},
url = {https://proceedings.neurips.cc/paper_files/paper/2022/file/cf4c7ee0734cdfe09a099cf6cd7b117a-Paper-Conference.pdf},
volume = {35},
year = {2022}
}
Start by installing the required modules:
pip install -r requirements.txt
The dataset is available for download at the following link: magnet dataset and contains two folders: 1d and 2d for the 1D and 2D PDE datasets respectively.
The structure of the 1D dataset is as follows:
├───E1
│ ├───irregular
│ │ CE_test_E1_graph_100.h5
│ │ CE_test_E1_graph_200.h5
│ │ CE_test_E1_graph_40.h5
│ │ CE_test_E1_graph_50.h5
│ │ CE_train_E1_graph_30.h5
│ │ CE_train_E1_graph_50.h5
│ │ CE_train_E1_graph_70.h5
│ │
│ └───regular
│ CE_test_E1_100.h5
│ CE_test_E1_200.h5
│ CE_test_E1_40.h5
│ CE_test_E1_50.h5
│ CE_train_E1_50.h5
│
├───E2
│ └───regular
│ CE_train_E2_50.h5
│ CE_test_E2_100.h5
│ CE_test_E2_200.h5
│ CE_test_E2_40.h5
│ CE_test_E2_50.h5
│
└───E3
└───regular
CE_test_E3_100.h5
CE_test_E3_200.h5
CE_test_E3_40.h5
CE_test_E3_50.h5
CE_train_E3_50.h5
Each file is formatted as follows: CE_{mode}_{dataset}_{resolution}.h5 where mode can be train or test and dataset can be E1, E2 or E3 and resolution denotes the resolution of the dataset. The folder regular contains simulations on a regular grid and irregular contains simulations on an irregular grid.
For the 2D dataset, it is structured as follows:
├── B1
│ ├── burgers_test_B1_128.h5
│ ├── burgers_test_B1_256.h5
│ ├── burgers_test_B1_32.h5
│ ├── burgers_test_B1_64.h5
│ ├── burgers_train_B1_128.h5
│ ├── burgers_train_B1_256.h5
│ ├── burgers_train_B1_32.h5
│ ├── burgers_train_B1_64.h5
│ ├── concentrated
│ │ ├── burgers_train_irregular_B1_128.h5
│ │ ├── burgers_train_irregular_B1_256.h5
│ │ ├── burgers_train_irregular_B1_512.h5
│ │ └── burgers_train_irregular_B1_64.h5
│ └── uniform
│ ├── burgers_train_irregular_B1_128.h5
│ ├── burgers_train_irregular_B1_256.h5
│ ├── burgers_train_irregular_B1_512.h5
│ └── burgers_train_irregular_B1_64.h5
└── B2
├── burgers_test_B2_128.h5
├── burgers_test_B2_256.h5
├── burgers_test_B2_32.h5
├── burgers_test_B2_64.h5
├── burgers_train_B2_128.h5
├── burgers_train_B2_256.h5
├── burgers_train_B2_32.h5
└── burgers_train_B2_64.h5
Each file is formatted as follows: burgers_{mode}_{dataset}_{resolution}.h5 where mode can be train or test and dataset can be B1 or B2 and resolution is the resolution of the dataset. The folder concentrated contains simulations on an irregular grid where points are sampled around a region in the grid while uniform contains simulations on a uniform irregular grid.
We use hydra for config management and command line parsing so it's straightforward to run experiments using our code-base. Below is an example command for training the MAgNet[CNN] model on the E1 dataset for 250 epochs on four GPUs:
python run.py \
model=magnet_cnn \
name=magnet_cnn \
datamodule=h5_datamodule_implicit \
datamodule.train_path={train_path} \
datamodule.val_path={val_path}' \
datamodule.test_path={test_path} \
datamodule.nt_train=250 \
datamodule.nx_train={train_resolution} \
datamodule.nt_val=250 \
datamodule.nx_val={val_resolution} \
datamodule.nt_test=250 \
datamodule.nx_test={test_resolution} \
datamodule.samples=16 \
model.params.time_slice=25 \
trainer.max_epochs=250 \
trainer.gpus=4 \
trainer.strategy='ddp'
You can find the relevant scripts that were used to run experiments under the scripts folder.