This application helps to find whether the substring exists in the predefined string. This is often used to identify whether a short DNA sequence is present in some dataset of long DNA sequence. The Naive Algorithm would run in O(n^2) time in order to search for the substring. This Desktop application helps to find the substring in the text in O(n) time, because it uses the Karp-Rabin algorithm.
The Karp-Rabin algorithm aims to use skillful modulo and arithmetic operations as well as hashing functions to decrease the time complexity of matching exact substring of the text. Here is the application in Genetics: Looking for DNA sequnces in the precalculated Data Set of DNA sequence
- Java 17
- JavaFX.controls
- JavaFX.fxml
- The Application is build by using Maven
- Open a new IntelliJ project.
- Then, Git clone this repo to your project
https://github.com/ilkhom19/dna_matching
- Then Run the Driver.java file
- Can recieve up to 500,000 character DNA sequence.
- Computes the Hash value of each substring with Length "L"
- Can compute the search in less than a second for any input given( also depends on the machine )
- Interactive search animations
- Optimize the Hashing Function
- A feature to generate randomly distributed DNA sequences similar to real DNA's
-
The first field of the application recieves the long DNA Sequence.The DNA Sequence cannot be empty.
-
The next 2 fields recieves two integers 1 <= L <= 1000 and 1 <= N <= 1000 representing the length of each query and the number of short DNA queries in the predefined set.
-
In the last TextArea, each of the next N lines contains a DNA sequence consisting of exactly L characters.
-
The base DNA Sequence:
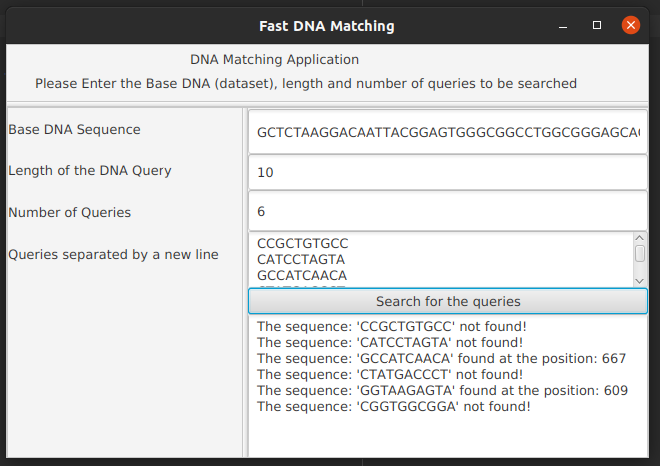
GCTCTAAGGACAATTACGGAGTGGGCGGCCTGGCGGGAGCACTACCCCATCGACGCGTACTCGAATACTGTATATTGCTCTCACATGAACAAATTAGTAGAGTGCCGCTTTCAGCCCCCCTGTCGTCGGCGACGTCTGTAAAATGGCGTTGATGTGGATCGACTCTATAGAGGCATCTACTGATGCGTAGGGAGATCCGGAATGTATTGGCCTATGTCACTGAAACTGTCCAAACACCCCATGTCGTTACTGAACGTATCGACGCATACCTCCTTCGTTGAGAACTCACAATTATACAACTGGGGACATAATCCCTACGCCCATCTTCTACACCCGTCTCTGTGGGTCCAGTTCAAGTGCTGGGAGAGCATCCTCCACAAGGTCTAGTGGTATGGTGGTATAGTAAGCTCGTACTGTGATACATGCGACAGGGGTAAGACCATCAGTAGTAGGGATAGTGCCAAAGCTCACTCACCACTGCCTATAAGGGGTGCTTACCTCTAGAATAAGTGTCAGCCAGTATAACCCCATGAGGAACCGAAAAGGCGAACCGGGCCAGACAATCCGGAGGCACCGGGCTCAAAGCCGCGACACGACGGCTCACAGCCGGTAAGAGTAACCCCGGAGTGAACACCTATGGGGCTGGATAAAACTGCCCTGGTGACCGCCATCAACAACCCGAATACGTGGCATTTCAGGAGGCGGCCGGAGGGGGGATGTTTTCTACTATTCGAGGCCGTTCGTTATAACTTGTTGCGTTCCTAGCCGCTATATTTGTCTCTTTGCCGACTAATGAGAACAACCACACCATAGCGATTTGACGCAGCGCCTCGGAATACCGTATCAGCAGGCGCCTCGTAAGGCCATTGCGAATACCAGGTATCGTGTAAGTAGCGTAGGCCCGTACGCGAGATAAACTGCTAGGGAACCGCGTCTCTACGACCGGTGCTCGATTTAATTTCGCCGACGTGATGACATTCCAGGCAGTGCCTCTGCCGCC
Lenth of the DNA Query: 10
Number of Queries: 6
CCGCTGTGCC
CATCCTAGTA
GCCATCAACA
CTATGACCCT
GGTAAGAGTA
CGGTGGCGGA
The sequence: 'CCGCTGTGCC' not found
The sequence: 'CATCCTAGTA' not found
The sequence: 'GCCATCAACA' found at the position: 667
The sequence: 'CTATGACCCT' not found
The sequence: 'GGTAAGAGTA' found at the position: 609
The sequence: 'CGGTGGCGGA' not found