HiGNN is a well-designed hierarchical and interactive informative graph neural networks framework for predicting molecular property by utilizing a co-representation learning of molecular graphs and chemically synthesizable BRICS fragments. Meanwhile, a plug-and-play feature-wise attention block was first designed in HiGNN architecture to adaptively recalibrate atomic features after message passing phase. HiGNN has been accepted for publication in Journal of Chemical Information and Modeling.
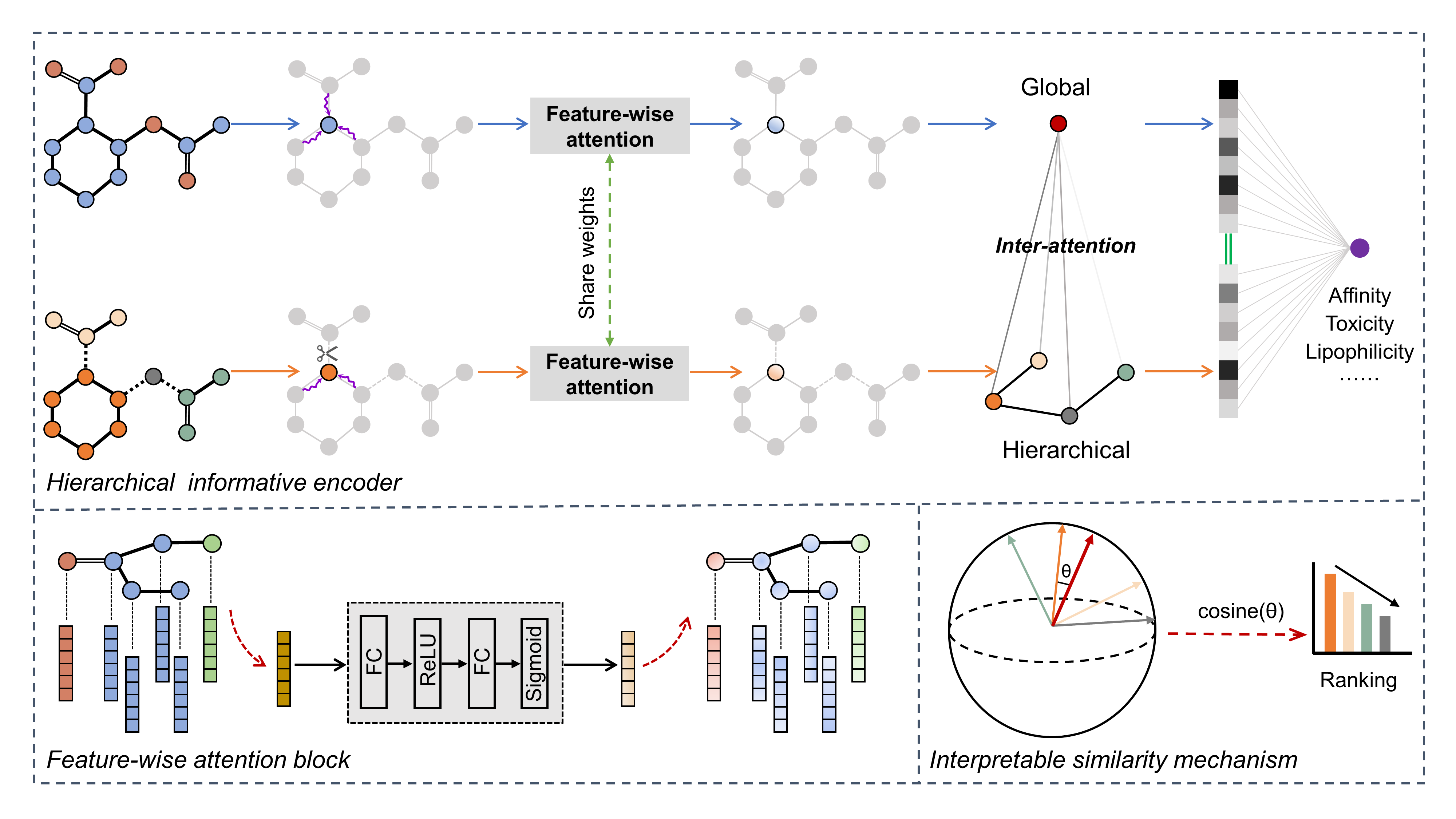 Fig.1 The overview of HiGNN
Fig.1 The overview of HiGNN
This project is developed using python 3.7.10, and mainly requires the following libraries.
rdkit==2021.03.1
scikit_learn==1.1.1
torch==1.7.1+cu101
torch_geometric==1.7.1
torch_scatter==2.0.7To install requirements:
pip install -r requirements.txtsource:the source code for HiGNN.config.pydatasets.pyutils.pymodel.pyloss.pytrain.pycross_validate.py
configs:HiGNN used yacs for experimental configuration, where you can customize the relevant hyperparameters for each experiment with yaml file.data:the dataset for training.raw:where to store the original csv dataset.processed:the dataset objects generated by PyG.
test:where the training logs, checkpoints and tensorboards are saved.example:jupyter notebook codes for fragments counting, t-SNE visualization, interpretation and so on.dataset:11 real-world drug-discovery-related datasets used in this study.
Taking the BBBP dataset as an example, experiment can be run via:
git clone https://github.com/idruglab/hignn
cd ./hignn
# For one random seed
python ./source/train.py --cfg ./configs/bbbp/bbbp.yaml --opts 'SEED' 2022 'MODEL.BRICS' True 'MODEL.F_ATT' True --tag seed_2022
# For 10 different random seeds (2021~2030)
python ./source/cross_validate.py --cfg ./configs/bbbp/bbbp.yaml --opts 'MODEL.BRICS' True 'MODEL.F_ATT' True 'HYPER' False --tag 10_seeds
# For hyperparameters optimization
python ./source/cross_validate.py --cfg ./configs/bbbp/bbbp.yaml --opts 'MODEL.BRICS' True 'MODEL.F_ATT' True --tag hignn # HiGNN
python ./source/cross_validate.py --cfg ./configs/bbbp/bbbp.yaml --opts 'MODEL.F_ATT' True --tag w/o_hi # the variant (w/o HI)
python ./source/cross_validate.py --cfg ./configs/bbbp/bbbp.yaml --opts 'MODEL.BRICS' True --tag w/o_fa # the variant (w/o FA)
python ./source/cross_validate.py --cfg ./configs/bbbp/bbbp.yaml --tag vanilla # the variant (w/o All)And more hyperparameter details can be found in config.py.
The interpretability of HiGNN can refer to interpretation_bace and interpretation_bbbp.
- The datasets used in this study are available in dataset or MoleculeNet.
- The training logs, checkpoints, and tensorboards for each dataset can be found in BaiduNetdisk.
In the present study, we evaluated the proposed HiGNN model on 11 commonly used and publicly available drug discovery-related datasets from Wu et al., including classification and regression tasks. According to previous studies, 14 learning tasks were designed based on 11 benchmark datasets, including 11 classification tasks based random- and scaffold-splitting methods and three regression tasks based on random-splitting method.
Table 1 Predictive performance results of HiGNN on the drug discovery-related benchmark datasets.
| Dataset | Split Type | Metric | Chemprop | GCN | GAT | Attentive FP | HRGCN+ | XGBoost | HiGNN |
|---|---|---|---|---|---|---|---|---|---|
| BACE | random | ROC-AUC | 0.898 | 0.898 | 0.886 | 0.876 | 0.891 | 0.889 | 0.890 |
| scaffold | ROC-AUC | 0.857 | 0.882 | ||||||
| HIV | random | ROC-AUC | 0.827 | 0.834 | 0.826 | 0.822 | 0.824 | 0.816 | 0.816 |
| scaffold | ROC-AUC | 0.794 | 0.802 | ||||||
| MUV | random | PRC-AUC | 0.053 | 0.061 | 0.057 | 0.038 | 0.082 | 0.068 | 0.186 |
| Tox21 | random | ROC-AUC | 0.854 | 0.836 | 0.835 | 0.852 | 0.848 | 0.836 | 0.856 |
| ToxCast | random | ROC-AUC | 0.764 | 0.770 | 0.768 | 0.794 | 0.793 | 0.774 | 0.781 |
| BBBP | random | ROC-AUC | 0.917 | 0.903 | 0.898 | 0.887 | 0.926 | 0.926 | 0.932 |
| scaffold | ROC-AUC | 0.886 | 0.927 | ||||||
| ClinTox | random | ROC-AUC | 0.897 | 0.895 | 0.888 | 0.904 | 0.899 | 0.911 | 0.930 |
| SIDER | random | ROC-AUC | 0.658 | 0.634 | 0.627 | 0.623 | 0.641 | 0.642 | 0.651 |
| FreeSolv | random | RMSE | 1.009 | 1.149 | 1.304 | 1.091 | 0.926 | 1.025 | 0.915 |
| ESOL | random | RMSE | 0.587 | 0.708 | 0.658 | 0.587 | 0.563 | 0.582 | 0.532 |
| Lipo | random | RMSE | 0.563 | 0.664 | 0.683 | 0.553 | 0.603 | 0.574 | 0.549 |
The code was partly built based on chemprop, TrimNet and Swin Transformer. Thanks a lot for their open source codes!