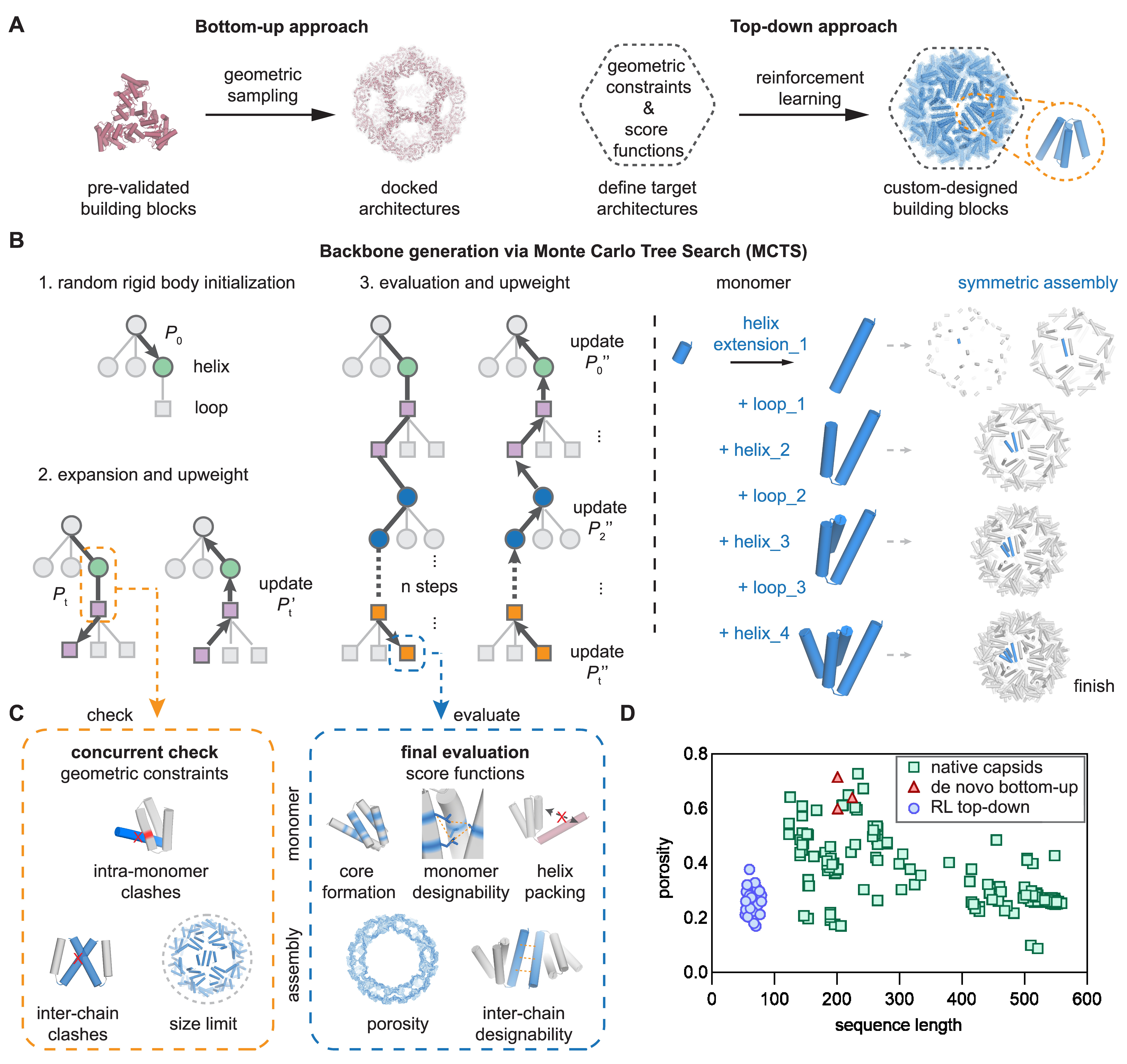
A generative protein design method for assembling alpha helices and loops into optimal protein backbones using provided sets of geometric constraints and score functions, as described in the paper "Top-down design of protein architectures with reinforcement learning".
Use this container (download size 4.6GB) to run the sampling scripts:
protein-backbone-MCTS.sif I_cages.py
Tetrahedral, octahedral, and icosahedral cages can be sampled as above without additional arguments, while the other scripts require additional arguments.
For shape filling, provide a .obj file specificying the desired shape:
protein-backbone-MCTS.sif fill_shapes.py example_shapes/test_sphere.obj
For C symmetries, provide a C symmetry number:
protein-backbone-MCTS.sif C_sym.py 8
For pore closure, a PDB is provided with docked outer and inner rings to initialize the sampling:
protein-backbone-MCTS.sif pore_closure.py example_pore_starts/sampled_HALC6_208_0_0_10.pdb
For quasisymmetric cages, provide a transformation matrices file specifying the desired symmetry:
protein-backbone-MCTS.sif quasisym_cages.py xforms/T2_7ncr.xforms
Sampled backbones passing filtering criteria are output in the outputs/ folder. The python scripts can be modified to customize the geometric constraints and score functions, sampling procedure and parameters, and final filtering criteria in order to generate desired backbones.
Zenodo archive can be found here:
Contact Isaac Lutz ([email protected], [email protected]) with any questions or comments.