Make sure the dependencies have been installed.
- cmake >= 3.10
- gcc >= 6.4
- zlib >= 1.2
Install CompSeed from source.
git clone https://github.com/i-xiaohu/CompSeed.git
cd CompSeed; mkdir build; cd build
cmake ..; make
If the installation is successful, the build subdirectory will contain the executable files.
bwaidxto create FM-index for a reference file.bwamemto run BWA-MEM (v0.7.17) for sequencing reads.CompSeedto run compressive seeding for reordered reads that generates same seeds and alignments as BWA-MEM.
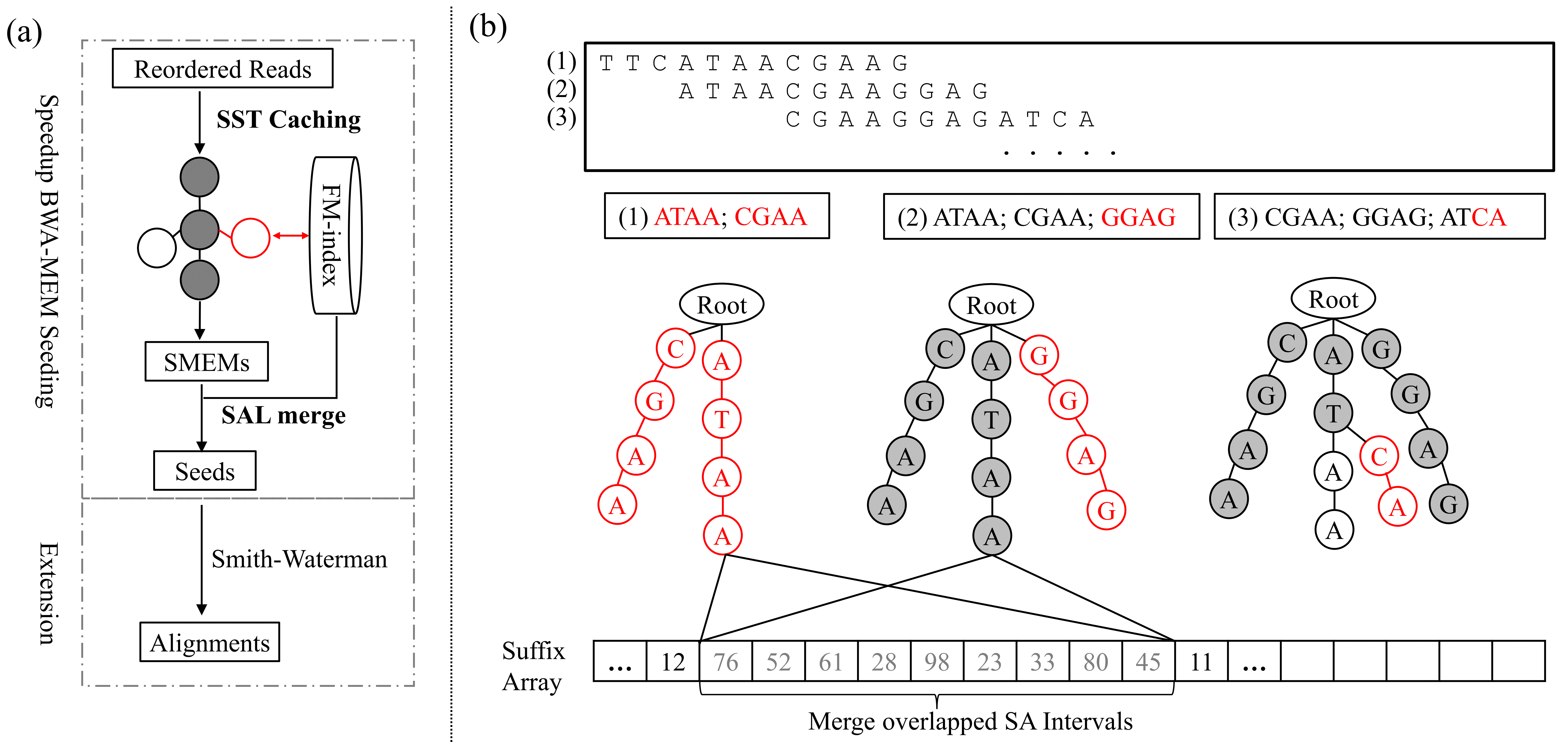
CompSeed is an algorithm demonstration for compressive alignment. It received the reads compressed and reordered by upstream reordering-based compressors, including SPRING, Minicom and PgRC. While CompSeed currently only supporst for single-end compression and alignment, the project of integrating compression and alignment is underway.
Build the FM-index for the reference sequence, for example hg19.fna.
bwaidx -p hg19 hg19.fnaCompress sequencing data with reordering-based compressors.
spring -c -t 16 --no-ids --no-quality -r -i data.fq -o data.spring
minicom -r data.fq -t 16; mv data_comp.minicom data.minicom
pgrc -t 16 -i data.fq data.pgrcDecompress to obtain the reordered reads.
spring -d -t 16 -i data.spring -o spring.reads
minicom -d data.mincom -t 16; mv data_dec.reads minicom.reads
pgrc -t 16 -d data.pgrc; mv data.pgrc_out pgrc.readsRun BWA-MEM.
bwamem -t 16 hg19 data.fq > bwa.samRun CompSeed.
CompSeed -t 16 hg19 spring.reads > css.sam
CompSeed -t 16 hg19 minicom.reads > csm.sam
CompSeed -t 16 hg19 pgrc.reads > csp.samFor CompSeed, all the original parameters of BWA-MEM seeding are supported.
-t number of threads
-k minimum seed length
-r look for internal seeds inside a seed longer than {-k} * {-r}
-y seed occurrence for the 3rd round seeding
-c skip seeds with more than {-c} occurrences
-K process {-K} input bases in each batch regardless of nThreads (for reproducibility)
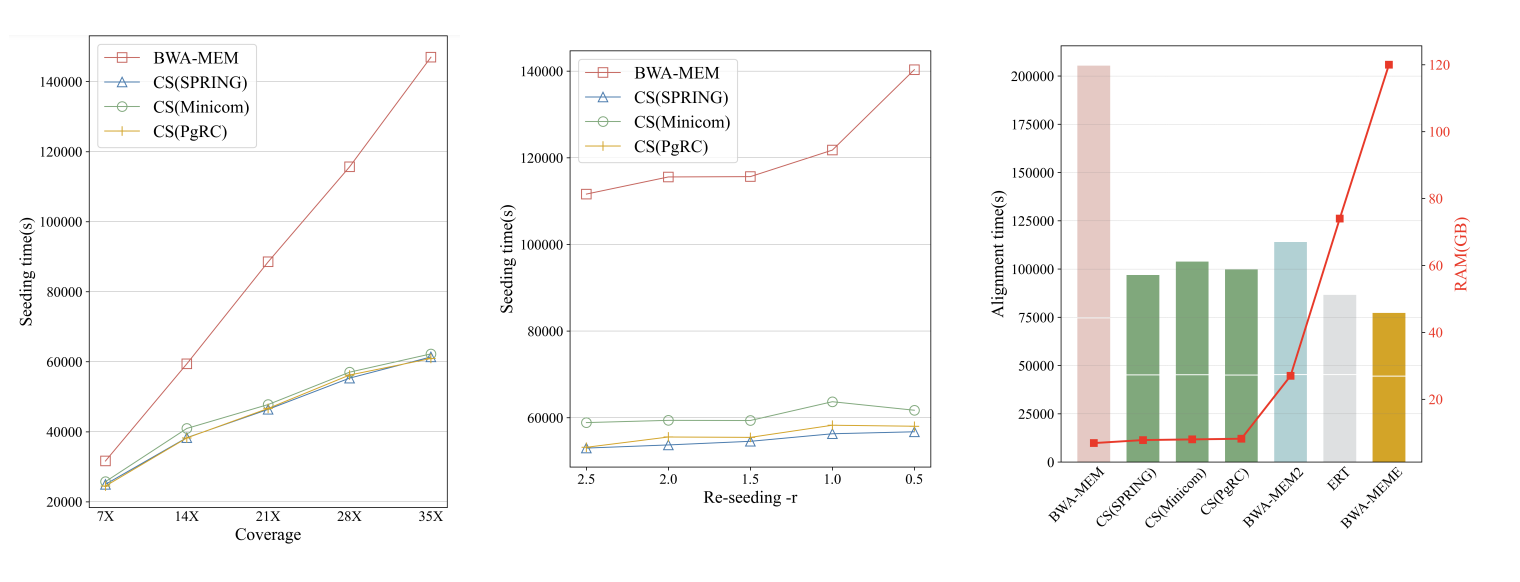
CompSeed fully utilizes the redundancy information provided from upstream compressors using trie structures, and avoids ~50% of the redundant time-consuming FM-index operations during the BWA-MEM seeding process.
After combined with AVX instructions for extension stage, a doubled alignment throughput is observed.
It shows enhanced performance as sequencing coverage increases, and it is almost not affected by the re-seeding parameter. Moreover, it has substantial memory advantage compared with the existing solutions, because it does not replace or modify the FM-index. All the acceleration benefits from the compression, thus does not conflict with existing hardware-based optimizations.
- Chandak, S., et al. (2019) SPRING: a next-generation compressor for FASTQ data, Bioinformatics, 35, 2674-2676.
- Liu, Y., et al. (2019) Index suffix–prefix overlaps by (w, k)-minimizer to generate long contigs for reads compression, Bioinformatics, 35, 2066-2074.
- Kowalski, T.M. and Grabowski, S. (2020) PgRC: pseudogenome-based read compressor, Bioinformatics, 36, 2082-2089.
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM.