⚠️ This work is in early development and changing rapidly. It is not ready for scientific use.⚠️
We hypothesize the process of science stands to benefit from having the option to suddenly become interactive and shareable - allowing for the poking or plucking, pushing or pulling, drilling in or out, grouping or separating, and sending or receiving of what would otherwise be a static snapshot of the data. The combined use of HoloViz and Bokeh tools could provide the interactivity, shareability, and scalability needed to support research as a collective action rather than a collection of solitary observations.
One of our overall goals is to facilitate the creation of fully open, reproducible, OS-independent, browser-based workflows for biomedical research primarily using sustainable, domain-independent visualization tools. In support of this goal, this repository is the development ground for optimization of HoloViz and Bokeh tools within the realm of neuroscience.
Specific repo objectives:
- Workflow Development: Host the development versions of workflows, facilitating consistency and code sharing across them.
- Collaboration Hub: Foster collaborative efforts between the developer teams and scientific collaborators outside these groups - aiming to effectively tailor development to specific requirements of the neuroscience community.
- Project Management: Track ideas, feedback, requirements, specifications, issues, requests, topic research, and progress in the associated Project Board and Meeting Notes.
- Host Domain-Specific Scripts: For instance, simulated data generators.
- Temporarily Host Benchmark Tooling: Eventually, to be be migrated to a dedicated, domain-independent repository.
This repository contains developmental versions of workflows, which can be loosely categorized into two types: generalized and specialized. Generalized workflows aim to be broadly applicable and primarily utilize domain-independent Pandata tools such as Numpy, Pandas, Xarray, SciPy, etc. These generalized workflows serve as the foundational building blocks for specialized workflows. Specialized workflows are designed to cater to specific contexts and have no limitations on the use of domain-specific tools.
| Title | Example Modality | Thumbnail | Info & Links | Description |
|---|---|---|---|---|
| Multi-Channel Timeseries | eeg, ephys | 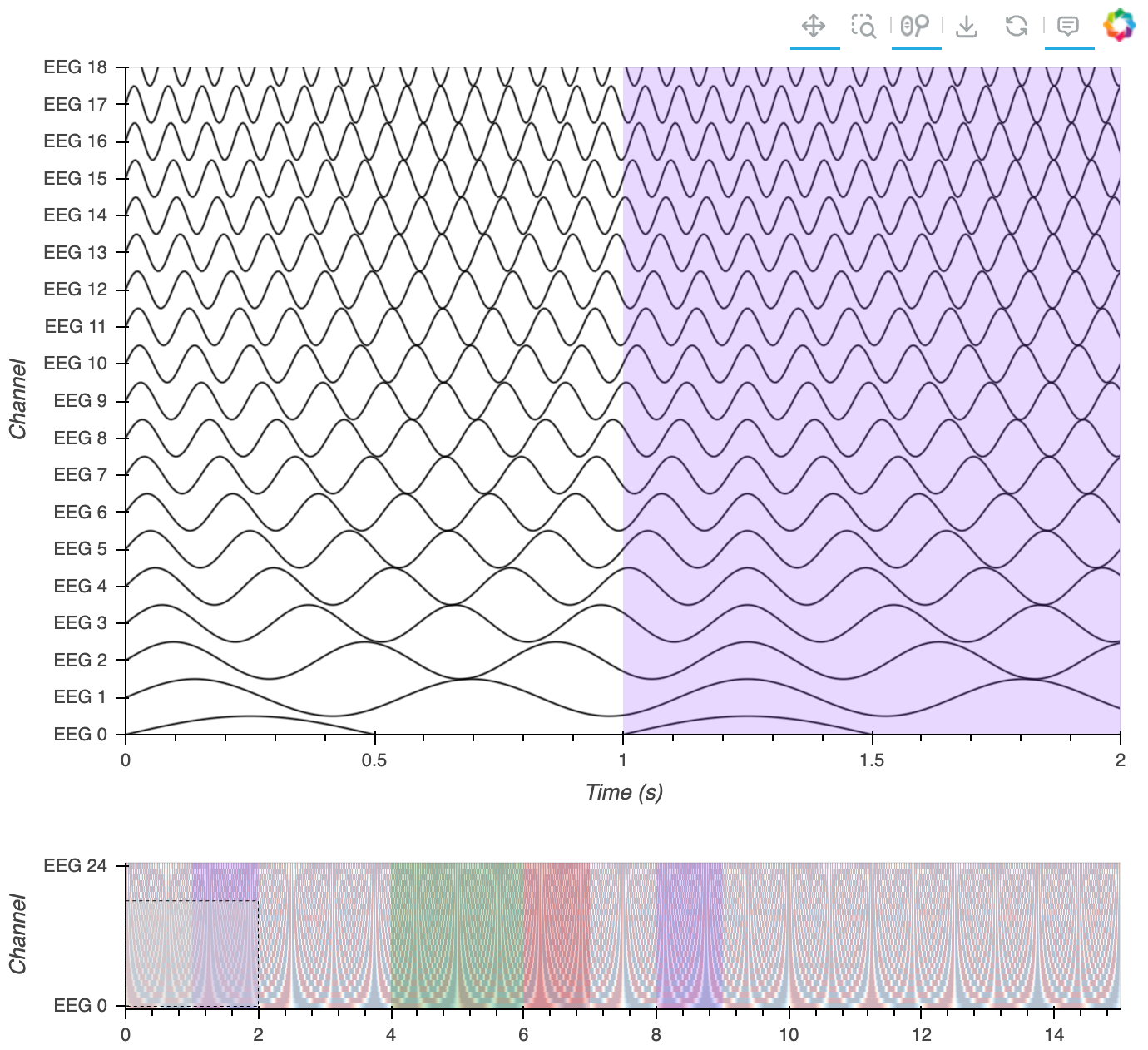 |
 workflow |
Synchronized examination of stacked time-series with large data handling, scale bar, annotations, minimap, and signal grouping. |
| Deep Image Stack | miniscope imaging | 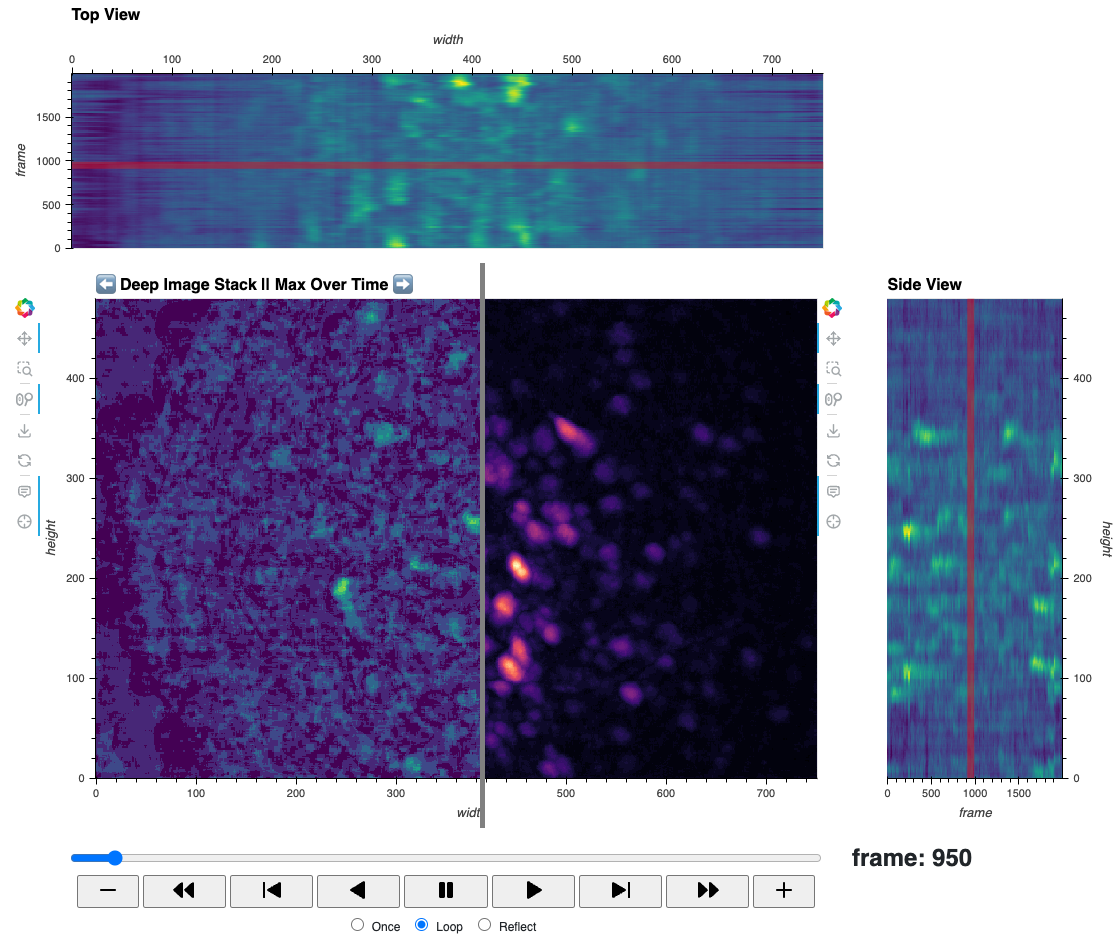 |
 workflow |
Efficient visualization of deep 2D calcium imaging movies with, playback controls, 2D annotation, scale bar, time views, intensity histogram, and summary statistics. |
| Waveform | ephys | 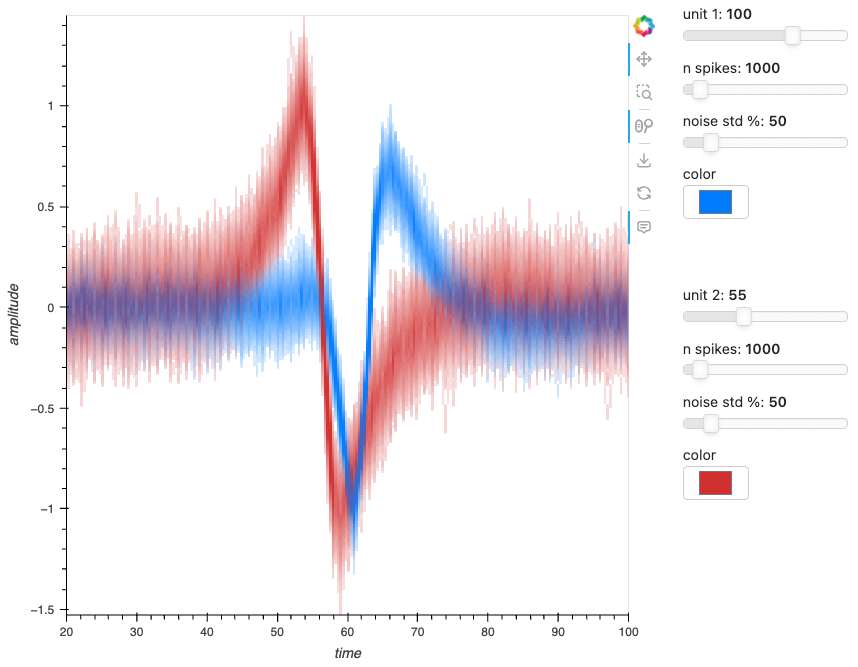 |
 workflow |
Oscilloscope-style display of action potential waveform snippets |
| Spike Raster | ephys | 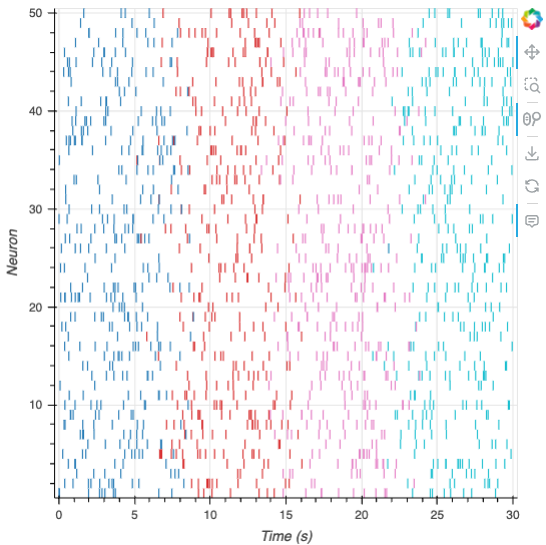 |
 workflow |
Efficient visualization of large-scale neuronal spike time data, with a simple API, aggregate views of spike counts, and spike-level metadata management |
Streaming data - extend the ephys, eeg, and/or video viewer workflows to display live streaming data.
Multimodal - visualizing and aligning ca-imaging with simultaneously recorded (but differently sampled) timeseries like EEG, EMG, and/or behavior. Alternatively, visualizing behavioral video (eye tracking, maze running) with timeseries data.
Linked electrode-array layout
| Title | Example Modality | Thumbnail | Info & Links | Description |
|---|---|---|---|---|
| Neuroglancer notebook | electron microscopy, histology | 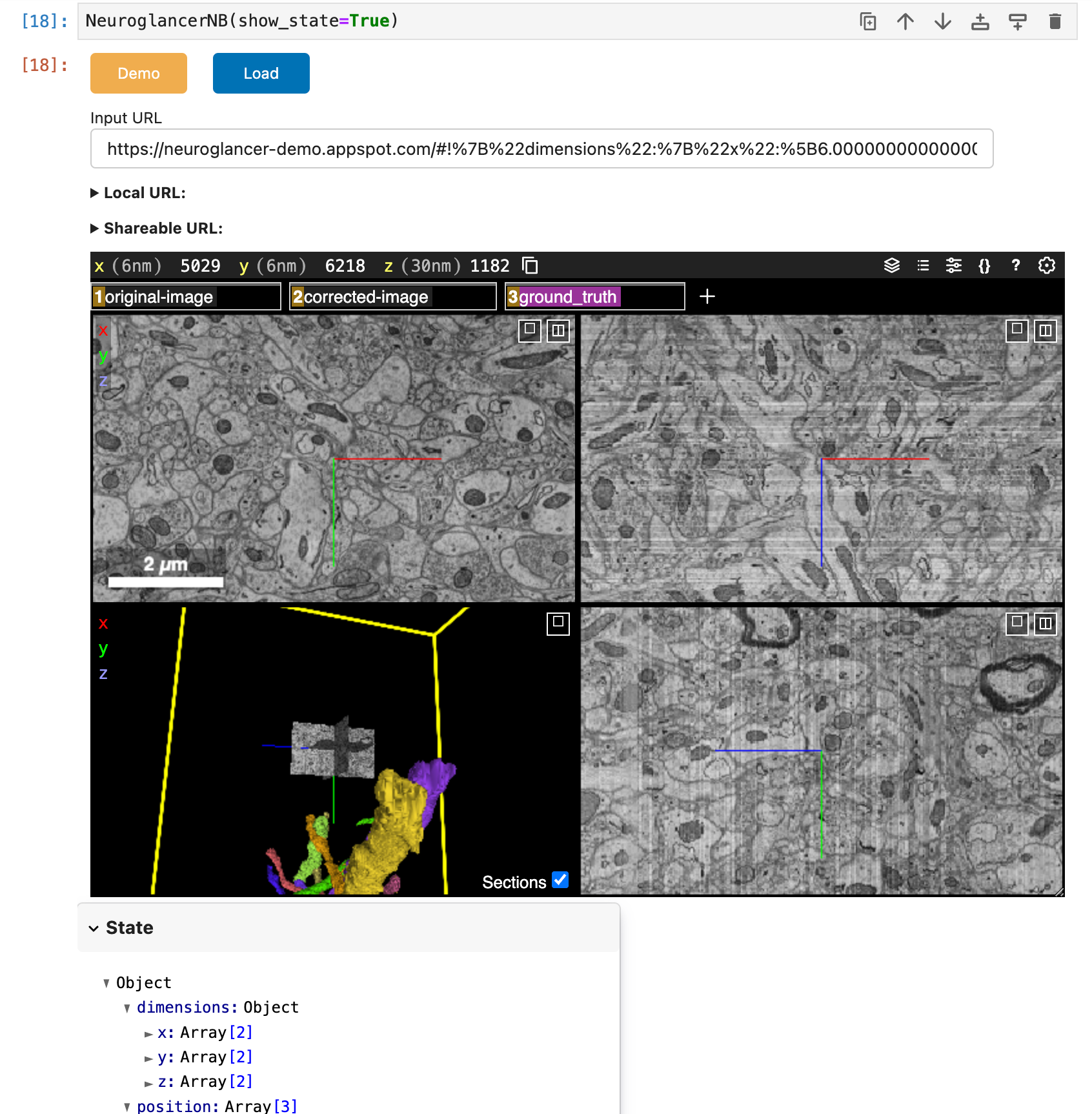 |
 workflow |
Notebook-based workflow for visualizing 3D volumetric data in a Neuroglancer application |
Spike Motif
MNE integration
Minian CNMF Temporal update parameter exploration app long timeseries improvement workflows/neuroglancer_notebook/assets/20240612_neuroglancerNB.png
- Workflows will be shared with the broader scientific community as they are ready. The target date for a first round of workflows is the end of 2024. Completed workflows will be listed on examples.holoviz.org, while select aspects will also go into the relevant Bokeh and HoloViz documentation pages.
- Workflow progress will be presented at the CZI open science conference in Boston, MA in June 2024.
- If you have ideas for where our workflows might be cross-linked of hosted, please reach out! We would love it if there was also a central place for bioscience workflows, like the Geoscience community has with Project Pythia.
- We are actively looking for opportunities to deliver tutorials, workshops, or other educational resources to help researchers in underrepresented communities effectively utilize our tools. Reach out on Discord if you want to brainstorm some ideas!
- Visit the Community page on HoloViz.org for more ways to join the conversation.
- If you want to contribute to the workflows or underlying libraries, read on for installation and contribution instructions.
This work is a collaboration between developers and scientists, and some developer-scientists. While some contributions are visible through the GitHub repo, many other contributions are less visible yet equally important.
Funding:
- 2023 - 2024: Chan Zuckerberg Initiative. Learn more in the grant announcement.
- Project Lead: Dr. Demetris Roumis (@droumis on Discord)
- HoloViz Director: Dr. James (Jim) Bednar (@jbednar on Discord)
- Bokeh Director: Bryan Van de Ven ([email protected])
Before installing the workflow environments, make sure you have Miniconda installed. If not, you can download and install it from the official site.
-
Clone the Repository: Clone the
neurorepository to your local machine.git clone https://github.com/holoviz-topics/neuro.git
-
Navigate to Workflow: Change to the directory of the workflow you're interested in.
cd neuro/workflows/<workflow>
-
Create Environment: Use
condato create a new environment from theenvironment.ymlfile.conda env create -f environment.yml
-
Activate Environment: After the environment is created, activate it.
conda activate <environment>
If you've already installed a workflow environment and the environment.yml file has been updated, follow these steps to update the environment:
-
Update Repository: Pull the latest changes from the repository.
git pull
-
Navigate to Workflow: Go to the directory of the workflow you're interested in.
cd neuro/workflows/<workflow>
-
Update Environment: Update the existing Conda environment based on the latest
environment.ymlfile.conda env update -f environment.yml --prune
The --prune option will remove packages from the environment not present in the updated environment.yml file.
- Task Management: As workflows are developed and honed, performance and UI bottlenecks will be identified and addressed. Some improvements for the workflows themselves will be within this repo, but many improvements will be in the appropriate underlying libraries within the HoloViz, Bokeh, or other GitHub Organizations. We will do our best to track the disparate tasks related to these efforts into this project board.
- Communication:
- Meeting minutes: Logged in the Wiki > Meeting Notes whenever possible.
- HoloViz Discord #neuro channel for real-time chat (if archived, post on the General HoloViz Discord channel)
- holoviz-topics/neuro GitHub repo issue tracker
- Specifications and Research: The Wiki has some data specifications and modality notes.
- Data Generation: To assist the development using real data, some workflows utilize simple data generators to help benchmark across data and parameter space. As the data generators/simulators can be useful to multiple workflows, they are kept as a separate module (
/src/neurodatagen). - Visualization source code: If there is visualization code or utilities that we want to live separate from the individual workflows, we can store them in
/src/hvneurofor now. However, this should be considered a temporary space until the code can be incorporated into existing libraries, or live in particular workflows. - Repo Structure and dev patterns:
/workflows /example1 readme_example1.md workflow_example1.ipynb environment.yml /dev date_example-workflow_task.ipynb- Use
readme_<workflow>.mdfor any essential workflow-specific info or links. - Maintain
workflow_<workflow>.ipynbas the latest version of the workflow. - Each workflow should have an
environment.ymlwith which to create a conda env - The
devdir in each workflow is scratch space. There is no expectation that anything here is maintained.
- Use

