- Fixed the bugs in batch_correct
- Add new functions to re-normalized inferred CSE
- Update new tutorial
User could install latest version ENIGMA through following command
install.packages("ENIGMA_v1.1.tar.gz",repos=NULL, type="source",INSTALL_opts=c("--no-multiarch"))
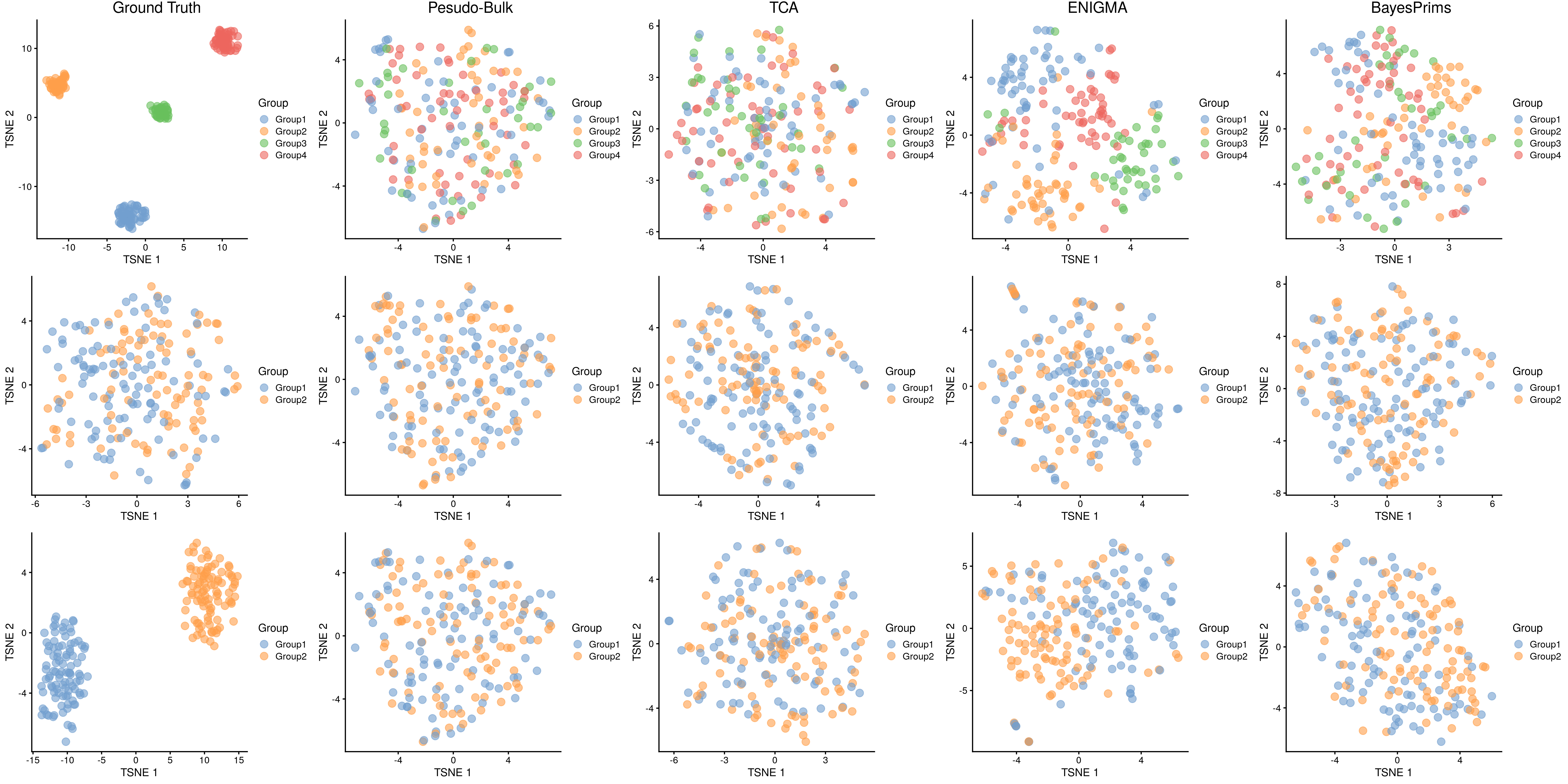
User could do benchmark through simulation, following this code to reproduce following figure
ENIGMA has three main steps. First, ENIGMA requires cell type reference expression matrix (signature matrix), which could be derived from either FACS RNA-seq or scRNA-seq datasets through calculating the average expression value of each gene from each cell type. Previous researches have widely used reference matrix curated from different platforms, for instance, Newman et al. used LM22 immune signature matrix which derived from microarray platform to deconvolute bulk RNA-seq dataset. However, we have to note that use references from different platforms would introduce unwanted batch effect between reference and bulk RNA-seq matrix, especially for the reference matrix derived from low coverage scRNA-seq dataset. To overcome this challenge, we used previously presented method that is specifically designed for correcting batch effect among bulk RNA-seq matrix and reference matrix. Second, ENIGMA applied robust linear regression model to estimate each cell type fractions among samples based on reference matrix derived from the first step. Third, based on reference matrix and cell type fraction matrix estimated from step 1 and step 2, ENIGMA applied constrained matrix completion algorithm to deconvolute bulk RNA-seq matrix into CSE on sample-level. In order to constraint the model complexity to prevent overfitting, we proposed to use two different norm penalty functions to regularize resulted CSE. Finally, the returned CSE could be used to identify cell type-specific DEG, visualize each gene’s expression pattern on the cell type-specific manifold space (e.g. t-SNE, UMAP), and build the cell type-specific co-expression network to identify modules that relevant to phenotypes of interest
For usage examples and guided walkthroughs, check the vignettes directory of the repo.
devtools::install_github("WWXKenmo/ENIGMA")
Please refer to the document of ENIGMA for detailed guidence using ENIGMA as a R package.
sva, mgcv, nlme, genefilter, BiocParallel, purrr, MASS, nnls
This is the alpha version of ENIGMA
the datasets could be downloaded from this repository. (master branch)
