A classification pipeline for motor imagery left or right hand movement, for the purposes of stroke rehabilitation. This was produced during the G.tec Spring School BCI Hackathon. Team members include Hadjer Benmeziane, Brier Rigby Dames, Ghinwa Masri, Vanessa Arteaga, Ernesto Alonso, Carolina Jiménez, Afrooz Seyedebrahimi, Holly Wilson.
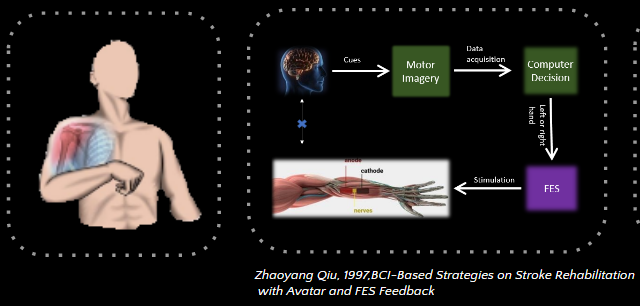
Stroke rehabilitation is an important topic, as stroke is the leading cause of serious and long-term disability worldwide. Yet, survivors can recover some motor function after rehabilitation therapy, for example using Functional Electrical Stimulation (FES). Our goal in this project, was to obtain EEG biomarkers from chronic stroke patients, to detect their intended movement of either their left or right hand.This can be useful to enable targeted improvement of their hand movements via for example FES.
We can frame this challenge as a classification problem, where we classify intended hand movement as left or right. Our aim was to improve upon a baseline classification accuracy given to us by the event organisers. The results given to us as a baseline to improve upon, used the traditional machine learning techniques, common spatial patterns with LDA, and time variant -LDA with PCA as seen in "Brain computer interface treatment for motor rehabilitation of upper extremity of stroke patients—A feasibility study" (Sebastián-Romagosa, Marc, et al., 2020).
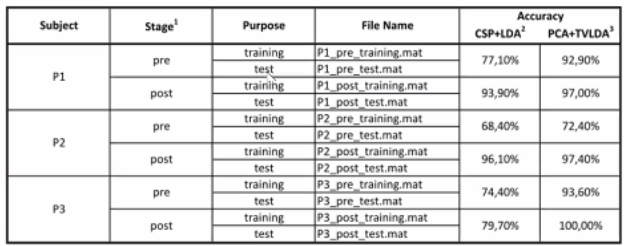
We obtained an EEG dataset of 3 chronic stroke patients, who performed a motor imagery task of either imagining moving their left or right hand when presented with a cue. The EEG data was gathered with a 16-channel cap, using 10/20 montage setup. The dataset is not publicly available and must be obtained directly from the authors. However for the purposes of our project, we had access to a .mat files, for each subject there were training and test data for pre and post FES stimulation (recorded in two separate sessions). Due to the non-stationarity of EEG data, we did not combined pre and post sessions within subjects and treated them separately.
We leveraged machine learning algorithms to extract the appropriate features and detect the intended movement. Our pipline includes two components: feature extraction, followed by classification.
For feature extraction, we experimented with the continuous wavelet transform. We chose to use wavelets, as opposed to fourier transform, as wavelets enables extraction and localization of transient and local components. By using wavelets we were able to obtain 32 characteristics for each channel, that well-defines the signals in both time and frequency domains.
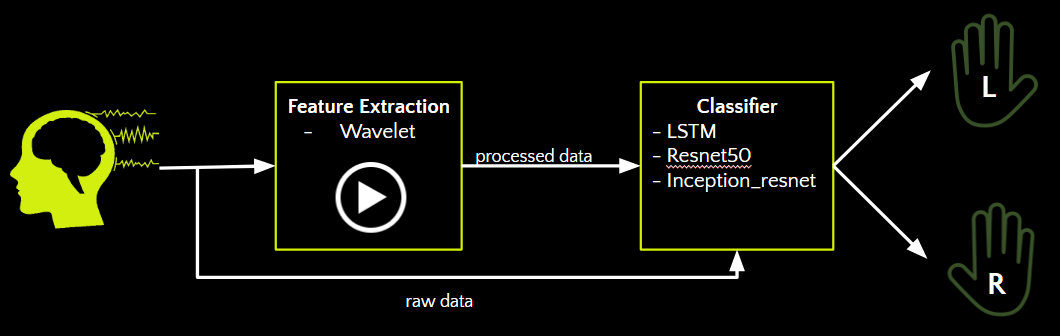
For classification, we leveraged traditional machine learning techniques, as well as deep learning classifiers. We experiment with using raw eeg data, and also with the wavelet eeg data as inputs.

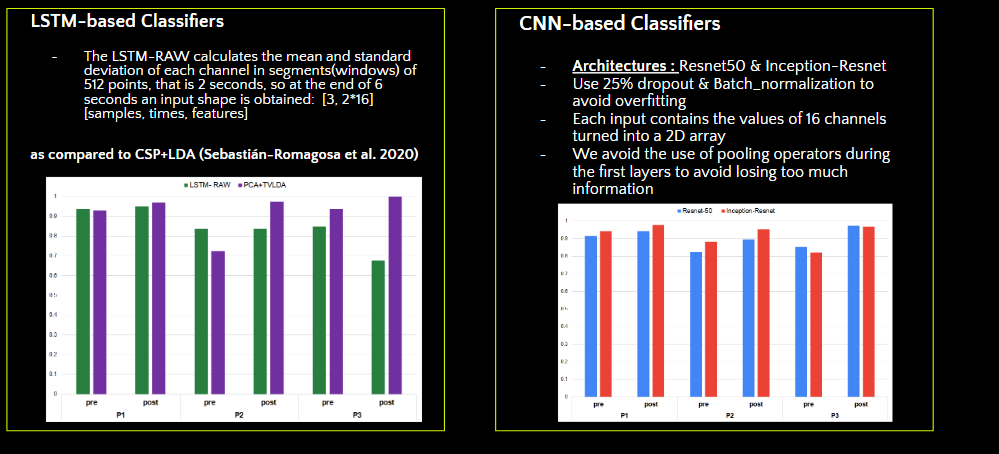
First, we considered the problem as a time-series classification, and implemented LSTMs, to exploit the temporal nature of EEG data. As you can see in the graph, using LSTMs, we got some promising results that we believe could be improved with more feature extraction and different RNN cells.
Next, we implemented CNNs. We used two standard CNN models, Resnet50 and Inception_resnet. Resnet50 uses special skip connections to deepen the model and extract more features, while inception_resnet improve resnet architecture by adding special blocks that include different types of convolutions.
In each, we omitted the first pooling layers to avoid losing too much information and included a dropout of 25%, to avoid overfitting as we have such a small amount of data per subjects. We first trained our models with all the training data for 30 epochs before fine-tuning again with the subjects data for 15 epochs. To adjust the models’ hyperparameters we used bayesian optimization.
We experimented with a range of traditional machine learning classifiers, including Ks Nearest Neighbour, Linear-Discriminant Analysis, Support Vector Machines, Random Forest and Logistic Regression.
Here we have a more detailed graph of some of our key results. Inception-Resnet had the best results, as shown in red. We found it obtained significantly higher accuracies than the baseline model common spatial patterns +LDA pipeline. Traditional ML techniques generally performed poorly, but we found the wavelet extraction boosted this performance.

If we had more time, we would like to have focused on making our classifiers more robust and generalisable, between individuals and also across sessions; collect more data or do some data augmentation on the channel level; compare CNN performance on raw with performance on the extracted features; focus on speed of classification for online-classification capabilities.