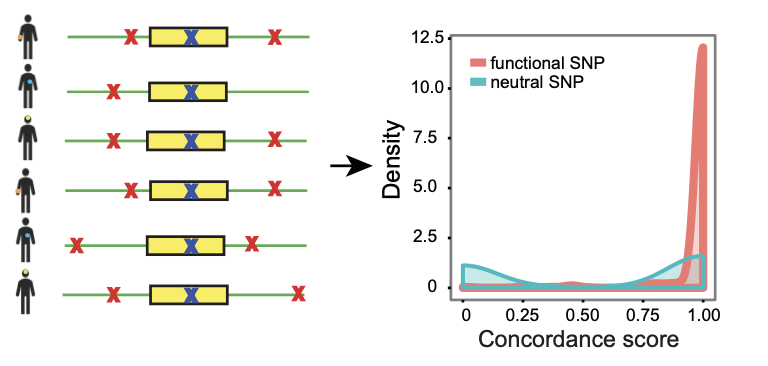
cGMAS (Concordance-based GMAS) is a method for predicting functional SNPs for GMAS (genetically-modulated alternative splicing) events. GMAS events have associated SNPs that can serve as tag SNPs. If a SNP is functional for GMAS, we expect to see a concordant SNP genotype and alternative splicing pattern accross a large number of individuals. We can quanitify the concordance between genotype and splicing pattern using a concordance score (Si). This method cannot distinguish true functional and neutal SNPs if they are in perfect LD. Also, the method requires a large number of individuals.
- Identify GMAS events (GMAS SNPs and GMAS exons)
- Determine genotype of candidate functional SNPs
B. Calculating concordance score (Si)
C. Filtering for candidate functional SNPs for GMAS
- Identify Si score peak
- Filter by mean Si scores and Si peak magnitude
- FDR corrections
We can use the ASARP pipeline to get high-quality tag SNPs and GMAS events as input for this pipeline.
Get the genotype of candidate SNPs. Possible sources: genotype information and RNA-seq data.
usage: gt.per.tissue.py [-h] -i infdir -a asasf -r refdir -t total -m mono -u
upperAR -l lowerAR -o outf
Get genotypes per tissue
optional arguments:
-h, --help show this help message and exit
-i infdir Input SNV count files dir
-a asasf Input asas file,asas events that are in at least X samples, sep by comma
-r refdir ref GT files directory
-t total total coverage to tell homozygous dbSNPs
-m mono mono coverage to tell homozygous dbSNPs
-u upperAR upper-bound allelic ratio for heterozygous dbSNPs (<=)
-l lowerAR lower-bound allelic ratio for heterozygous dbSNPs (>=)
-o outf Output file
Union of GMAS events from all samples in a study. It does NOT matter which individuals we identified the GMAS events from.
usage: candid.bed.py [-h] -i ind -d indir -s suff -m min -o outf
Get candidate functional SNPs
optional arguments:
-h, --help show this help message and exit
-i ind Individual of interest
-d indir Input file directory
-s suff file suffix
-m min min number of tissues to decide heterozygous
-o outf Output file
Each individual should have his or her specific list of SNPs and the genotype.
usage: tag.bed.py [-h] -i inf -a asasf -r ref -c cov -o outf
Get candidate functional SNP genotypes
optional arguments:
-h, --help show this help message and exit
-i inf Input candidate SNP bed file
-a asasf Input file with GMAS events that are in at least X samples
-r ref alleles count ref file
-c cov tag snv coverage
-o outf Output file
usage: splicing.concordance.py [-h] -i inf -d indiv -t tag -m maxD -o outf -a
anno -s search
Calculate concordance scores
optional arguments:
-h, --help show this help message and exit
-i inf Input candidate functional SNP bed file
-d indiv Input individual ID
-t tag tag snp bed file
-m maxD max d for RNA-seq defined tag snvs
-o outf Output file
-a anno gene annotation bed file
-s search max dist in nt from candidate functional snp to the AS exon to be
tested; input "INF" to test all possible snp pairs within the same
gene
1. Model Si scores of an event using GMM to get the Si score peak locations and to remove cases with shallow Si score peak magnitude.
usage: peak.si.rm.bg.py [-h] -i inf [-r ref] [-m min] -n N -o outf -b bin -p
het
Remove cases with shallow Si score peak magnitude
optional arguments:
-h, --help show this help message and exit
-i inf Input file prefix
-r ref SNVs to be filtered out
-m min min data points (individuals) in the Si distri
-n N Number of GMM components fitted
-o outf Output file
-b bin bin for randomization
-p het Percentage of heterozygous individuals
- v1
- number of individuals (n): 40
- P-value testing whether the GMM is significantly different from Si = 1 (p): 0.1
- Min Si (s): 0.8
- Min % individuals in major GMM (m): 0.9 -> applies to only tag SNVs that have high enough (n)
- Take out cases with all individuals who are homozygous (-M yes)
usage: get.causal.v1.py [-h] -i annoI -e annoE -r causalf -o outf -t tissue -s
si -p pval -n minPt -m major
Get functional SNPs - v1
optional arguments:
-h, --help show this help message and exit
-i annoI intron anno bed
-e annoE exon anno bed
-r functionalf ref functional si file dir
-o outf Output file
-t tissue tissue of interest
-s si min Si
-p pval min pval; pval is testing whether si is diff from 1
-n minPt min data points (indiv) per functional-exon-tag pair
-m major min membership ratio of the major component
- v2
- number of individuals (n): 40
- P-value testing whether the GMM is significantly different from Si = 1; Si = 0 (p): 0.1,0.01 -> >= 0.1 for Si = 1 and <= 0.01 for Si = 0
usage: get.causal.v2.py [-h] -i annoI -e annoE -r causalf -o outf -t tissue -p
pval -n minPt
Get functional SNPs - v2
optional arguments:
-h, --help show this help message and exit
-i annoI intron anno bed
-e annoE exon anno bed
-r functionalf ref functional si file dir
-o outf Output file
-t tissue tissue of interest
-p pval min pval; pval is testing whether si is diff from 1
-n minPt min data points (indiv) per functional-exon-tag pair
- v2b
- number of individuals (n): 40
- P-value testing whether the GMM is significantly different from Si = 1; Si = 0 (p): 0.05,0.05 -> <= 0.05 for Si = 1 and <= 0.05 for Si = 0
- Min % individuals in major GMM (m): 0.9 -> applies to only tag SNVs that have high enough (n)
- Min % het individuals (s; gtr): 0.95
usage: get.causal.v2b.py [-h] -i annoI -e annoE -r causalf -o outf -t tissue
-s gtr -p pval -n minPt -m major
Get functional SNPs - v2b
optional arguments:
-h, --help show this help message and exit
-i annoI intron anno bed
-e annoE exon anno bed
-r functionalf ref functional si file dir
-o outf Output file
-t tissue tissue of interest
-s gtr min GT ratio: RV/totalIndiv
-p pval min pval; pval is testing whether si is diff from 1
-n minPt min data points (indiv) per functional-exon-tag pair
-m major min membership ratio of the major component
- v1
usage: detect.non-causal.py [-h] -i inf [-r ref] -s suff -o outf
Detect non-functional SNPs - v1
optional arguments:
-h, --help show this help message and exit
-i inf Input file prefix
-r ref ref file with SNV tested
-s suff suff
-o outf Output file
- v2
usage: detect.non-causal.v2.py [-h] -i inf [-r ref] -s suff -o outf
Detect non-functional SNPs - v2
optional arguments:
-h, --help show this help message and exit
-i inf Input file prefix
-r ref ref file with SNV tested
-s suff suffix
-o outf Output file
- v2b
usage: detect.non-causal.v2b.py [-h] -i inf [-r ref] -s suff -o outf
Detect non-functional SNPs - v2b
optional arguments:
-h, --help show this help message and exit
-i inf Input file prefix
-r ref ref file with SNV tested
-s suff suffix
-o outf Output file
- v1 : fisherP.adjust.R (fisherP.adjust.sh)
- v2 : p.adjust.R (p0.adjust.sh)
- v2b: p.adjust.R (p0.adjust.v2b.sh)
- Esther Hsiao
- Grace Xiao lab