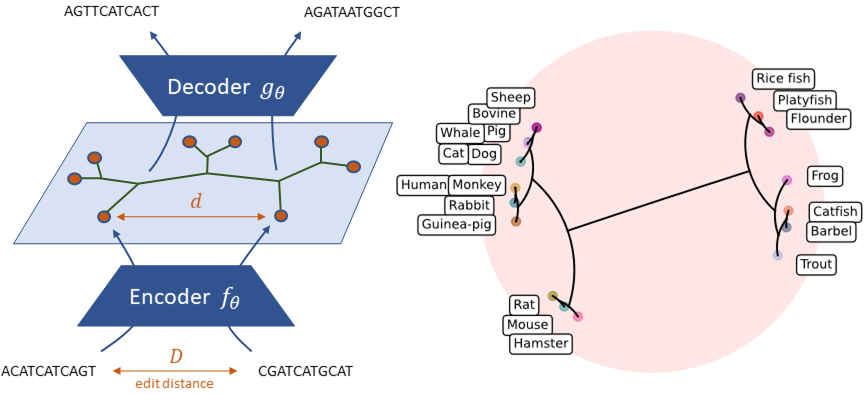
Official implementation of Neural Distance Embeddings for Biological Sequences (NeuroSEED) in PyTorch published at NeurIPS 2021 (preprint). NeuroSEED is a novel framework to embed biological sequences in geometric vector spaces.
Note: unfortunately due to my move between institutions the download scripts are broken and the files are no longer available on the original Drive. I have reuploaded them here, but reach out if you believe there are some missing files.
The repository is organised in four main folders one for each of the tasks analysed. Each of these contain scripts and models used for the task as well as instructions on how to run them and the tuned hyperparameters found.
edit_distancefor the edit distance approximation taskclosest_stringfor the closest string retrieval taskhierarchical_clusteringfor the hierarchical clustering task, further divided inrelaxedandunsupervisedfor the two approaches exploredmultiple_alignmentfor the multiple sequence alignment task, further divided inguide_treeandsteiner_stringutilcontains a series of utility routines shared between all the taskstestscontains a wide range of tests for the various components of the repository
Create a virtual (or conda) environment and install the dependencies:
python3 -m venv neuroseed
source neuroseed/bin/activate
pip install -r requirements.txt
Then install the mst and unionfind packages used for the hierarchical clustering:
cd hierarchical_clustering/relaxed/mst; python setup.py build_ext --inplace; cd ../../..
cd hierarchical_clustering/relaxed/unionfind; python setup.py build_ext --inplace; cd ../../..
@article{corso2021neuroseed,
title={Neural Distance Embeddings for Biological Sequences},
author={Corso, Gabriele and Ying, Rex and P{\'a}ndy, Michal and Veli{\v{c}}kovi{\'c}, Petar and Leskovec, Jure and Li{\`o}, Pietro},
journal={Advances in Neural Information Processing Systems},
year={2021}
}
MIT