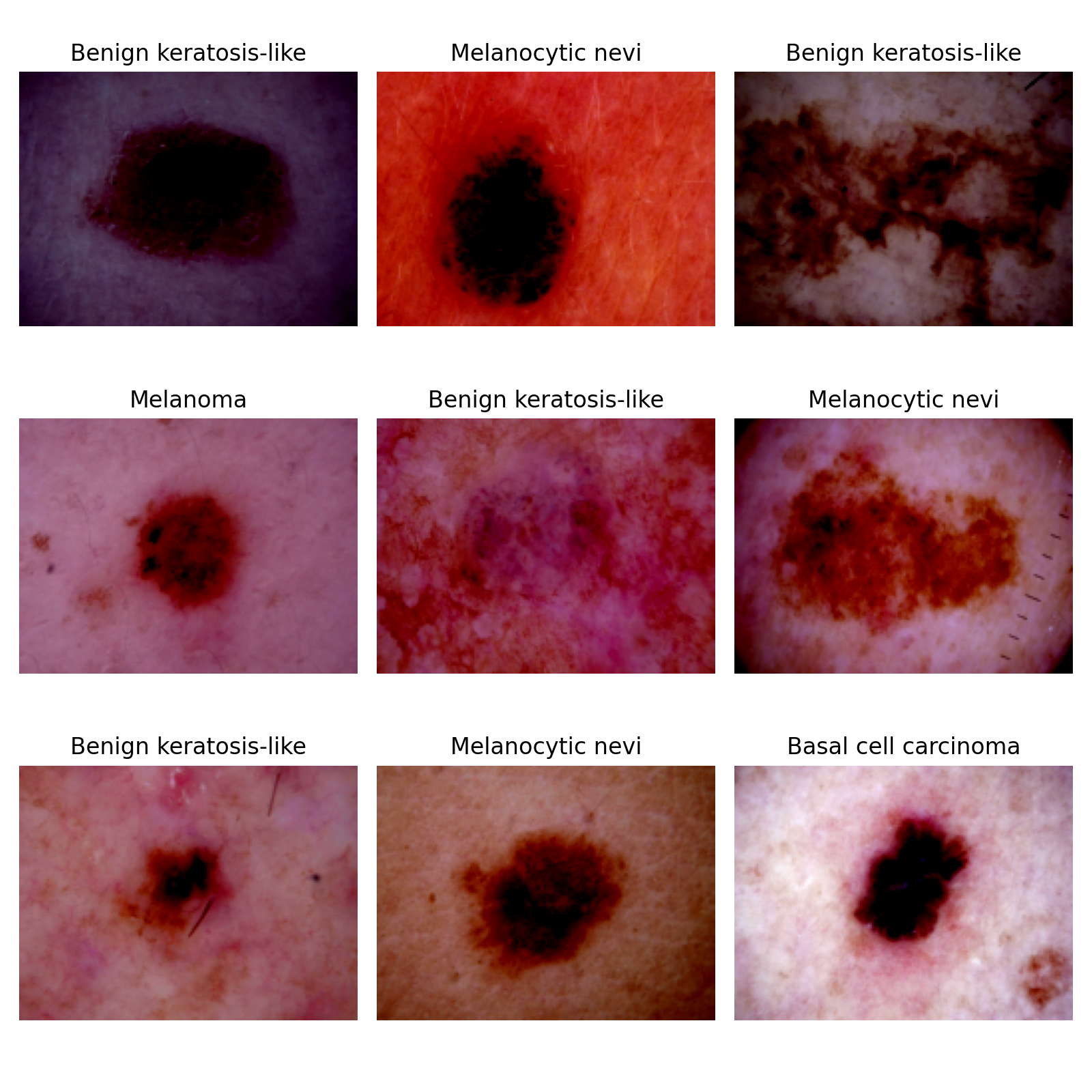
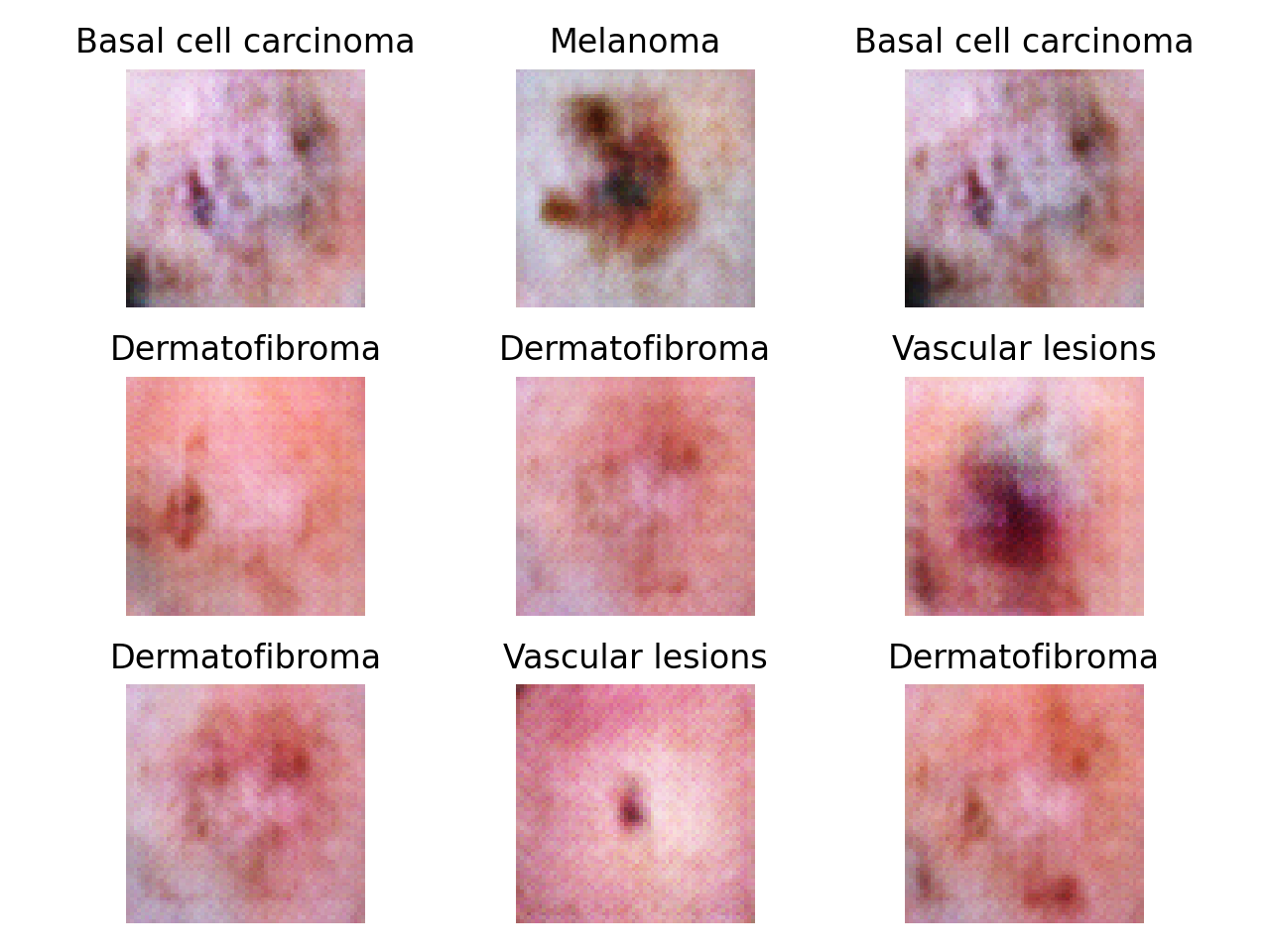
Automated diagnosis of any kind are hampered by the small size, lack of diversity and expensiveness of available dataset of medical images. To tackle this problem, several approaches using generative models have been applied.
Thence, in this project we will be using a (conditional) Deep Convolutional Generative Adversarial Network (cDCGAN) to synthetically generate medical images; concretely, dermatological images of pigmented skin lesions.
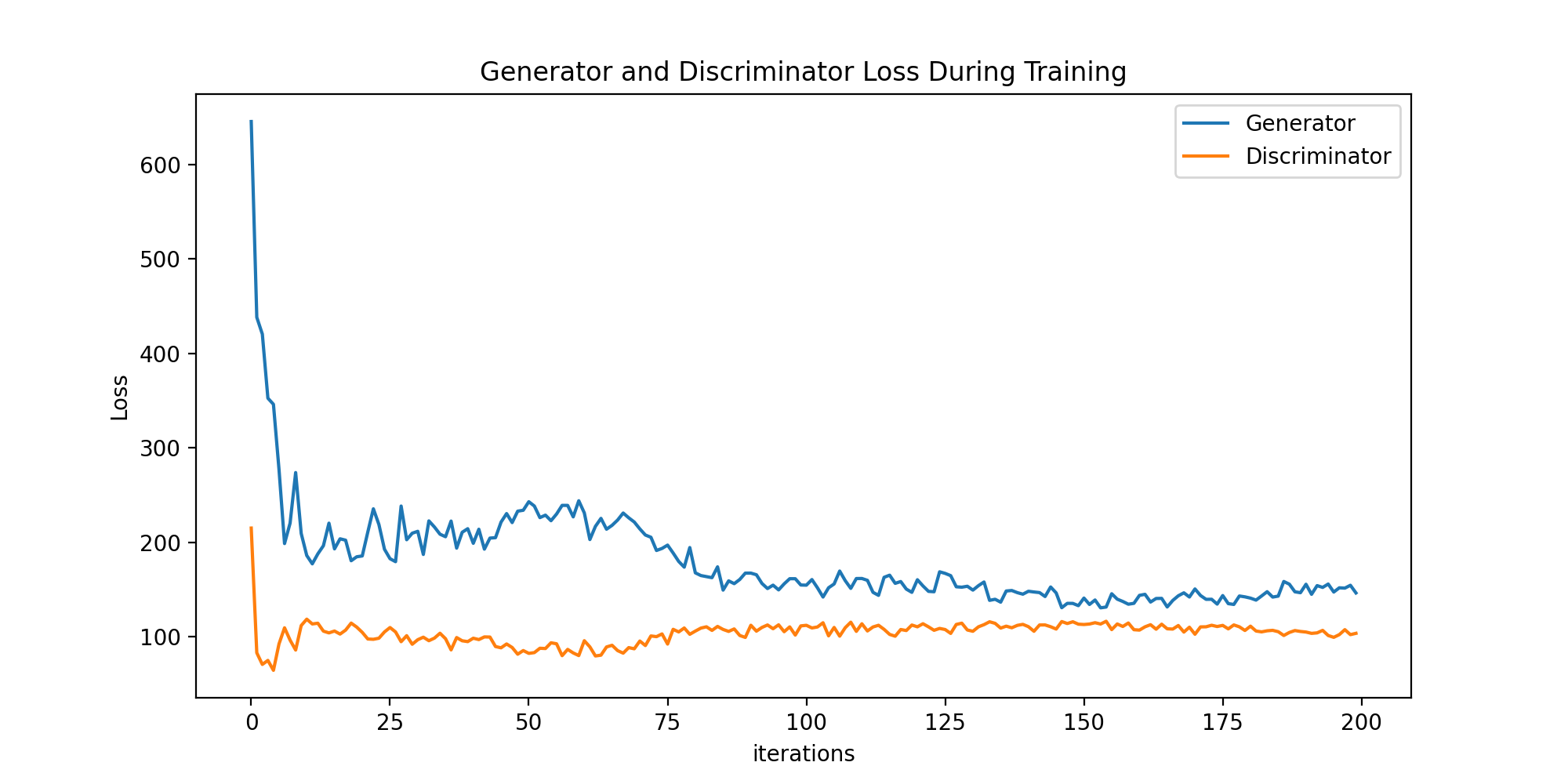
This were generated after a training with IMAGE_SIZE = 64 and the current
hyperparameters, which yielded the following error losses during training:
It is trivial to see the discriminator was too simple to keep improving at some point (its predictive performance was eventually at a standstill, preventing the generator to improve). This is due to the fact no fine-tuning was yet carried out.
We will use the HAM10000 dataset ("Human Against Machine with 10000 training images"), published in the 2018. In the words of the authors:
We collected dermatoscopic images from different populations acquired and stored by different modalities. Given this diversity we had to apply different acquisition and cleaning methods and developed semi-automatic workflows utilizing specifically trained neural networks. The final dataset consists of 11788 dermatoscopic images, of which 10010 will be released as a training set for academic machine learning purposes and will be publicly available through the ISIC archive. This benchmark dataset can be used for machine learning and for comparisons with human experts. Cases include a representative collection of all important diagnostic categories in the realm of pigmented lesions. More than 50% of lesions have been confirmed by pathology, the ground truth for the rest of the cases was either follow-up, expert consensus, or confirmation by in-vivo confocal microscopy.
This same dataset, has been uploaded at several places:
- Harvard Dataverse
- Kaggle: downloaded from here
Training of neural networks for automated diagnosis of pigmented skin lesions is hampered by the small size and lack of diversity of available dataset of dermatoscopic images. This problem was tackled by releasing the HAM10000 ("Human Against Machine with 10000 training images") dataset. The authors collected dermatoscopic images from different populations, acquired and stored by different modalities. The final dataset consists of 10015 dermatoscopic images which can serve as a training set for academic machine learning purposes. Cases include a representative collection of all important diagnostic categories in the realm of pigmented lesions:
- Actinic keratoses and intraepithelial carcinoma / Bowen's disease (
akiec,0labeled) - Basal cell carcinoma (
bcc,1labeled) - Benign keratosis-like lesions: solar lentigines / seborrheic keratoses and lichen-planus like keratoses (
bkl,2labeled) - Dermatofibroma (
df,3labeled) - Melanoma (
mel,4labeled) - Melanocytic nevi (
nv,5labeled) - Vascular lesions: angiomas, angiokeratomas, pyogenic granulomas and hemorrhage (
vasc,6labeled).
The ground-truth of the lesions are confirmed through:
- Histopathology (
histo): more than 50% of lesions - Follow-up examination (
follow_up) - Expert consensus (
consensus) - Confirmation by in-vivo confocal microscopy (
confocal). These are labeled accordingly in thedx_typecolumn of theHAM10000_metadata.csv.
Due to upload size limitations, images were stored in two files:
HAM10000_images_part_1: with 5000.jpgfilesHAM10000_images_part_2: with 5015.jpgfiles
The dataset includes lesions with multiple images, which can be tracked by the lesion_id column within the HAM10000_metadata.csv file. This file contains the following columns:
image_id:idand name with which the image can be found in one ofHAM10000_images_part_1orHAM10000_images_part_2folderslesion_id:idof the lesion (note one lesion can contain more than one image)dx_type: 4-category procedure of the diagnostic (through whichdxwas confirmed)age: age of the patient as integersex:maleorfemalein function of the biological sex of the patientlocalization: body part in which the lesion is founddx: the ground-truth, i.e. our label
There are plenty of medical images datasets. Below we list some websites which may prove useful:
- The MedMNIST v2 dataset is the MNIST of Medical Imaging containing a large-scale MNIST-like collection of standardized biomedical images.
- Torch's Medical Images Datasets
- Dermatology Image Bank from the University of Utah
There are lots of general datasets' repositories out there, which may be worht looking at:
To run the code locally (without any Docker container), I installed pytorch (with GPU support) in a dedicated conda environment by following this guide.
Note the cuDNN files have to be copied at
C:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8. Once, CUDA 11.8
and the compatible cuDNN version are installed, we set up GPU-pytorch
through conda according to the official website.
conda create -n hamgan python=3.9
conda activate hamgan
conda install pytorch torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia -y
conda install jupyter notebook pandas matplotlib seaborn -yIn order to run the repo .ipynb we need to set up our conda environment
as jupyter kernel, by typing:
python -m ipykernel install --user --name=hamgan