This repository hosts HINT, a deep learning based method for clinical trial outcome prediction. The repository can be mainly divided into three parts:
benchmarkdescribes the process of curating benchmark dataset named Trial Outcome Prediction (TOP) for clinical trial outcome prediction.HINTis the Hierarchical Interaction Network, a deep learning based method.datastores processed data.
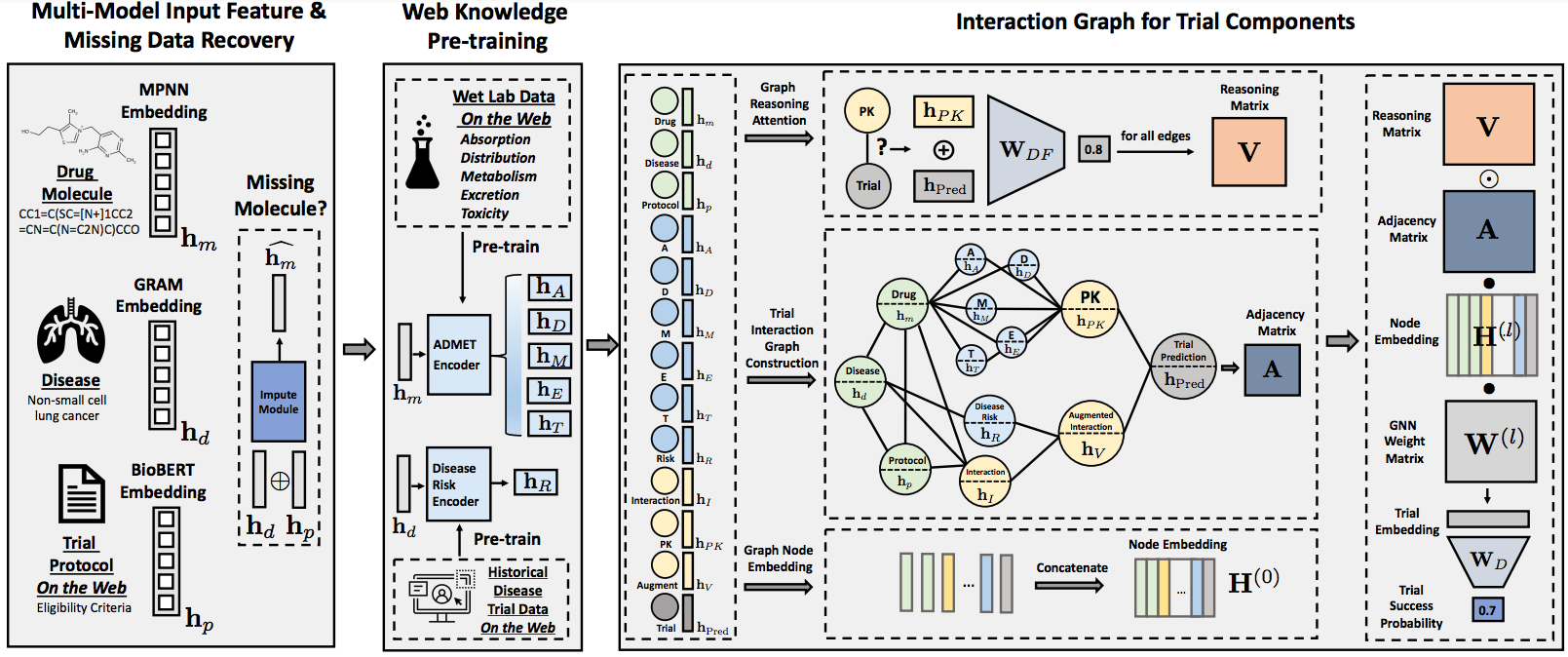
The following figure illustrates the pipeline of HINT.
We build conda environment and uses conda or pip to install the required packages. See conda.yml for all the packages.
conda create -n predict_drug_clinical_trial python==3.7
conda activate predict_drug_clinical_trial
conda install -c rdkit rdkit
pip install tqdm scikit-learn
pip install torch
pip install seaborn
pip install icd10-cmWe use following command to activate conda environment.
conda activate predict_drug_clinical_trialTo standardize the clinical trial outcome prediction, we create a benchmark dataset for Trial Outcome Prediction named TOP, which incorporate rich data components about clinical trials, including drug, disease and protocol (eligibility criteria).
All the scripts are in the folder benchmark.
Please see benchmark/README.md for details.
After processing the data, we learn the Hierarchical Interaction Network (HINT) on the following four tasks. The following figure illustrates the pipeline of HINT. All the scripts are available in the folder HINT.
Please see HINT/README.md for details.
We add the prediction results in ./results for all the three phases.
The trained HINT models for all the three phases are available in ./save_model.
benchmark:tutorial_benchmark.ipynbdescribes some key components of the data curation process.HINT:tutorial_HINT.ipynbis a tutorial to learn and evaluate HINT step by step.
Please contact [email protected] for help or submit an issue. This is a joint work with Kexin Huang, Cao(Danica) Xiao, Lucas M. Glass and Jimeng Sun.
The benchmark dataset and code (including data collection and preprocessing, model construction, learning process, evaluation), referred as the Works, are publicly available for Non-Commercial Use only at https://github.com/futianfan/clinical-trial-outcome-prediction. Non-Commercial Use is defined as for academic research or other non-profit educational use which is: (1) not-for-profit; (2) not conducted or funded (unless such funding confers no commercial rights to the funding entity) by an entity engaged in the commercial use, application or exploitation of works similar to the Works; and (3) not intended to produce works for commercial use.