A full-capture Hierarchical Bayesian model of Pollock's Closed Robust Design and application to dolphins
This contains a tutorial of several Robust Design (RD) Bayesian Mark-Recapture models, as presented in the paper Rankin RW, Nicholson K, Allen SJ, Krützen M, Bejder L, Pollock KH. 2016. A full-capture Hierarchical Bayesian model of Pollock's Closed Robust Design and application to dolphins. Frontiers in Marine Science 3:25, doi: 10.3389/fmars.2016.00025 (alt. download pdf here). This tutorial also uses data directly from the parent publication: Nicholson K, Bejder L, Allen SJ, Krützen M, and Pollock KH. 2012. Abundance, survival and temporary emigration of bottlenose dolphins (Tursiops sp.) off Useless Loop in the western gulf of Shark Bay, Western Australia. Marine and Freshwater Research 63:1059–1068. Please cite both these studies if using this code and data (See BIBTEX citations at the end of this read me).
The RD models are useful for the analysis of marked animals, estimating their abundance and demographic parameters, such as survival and movement. Importantly, the Hierarchical Bayesian (HB) model creates a flexible modelling environment to address several perennial issues encountered in Frequentist models 'fixed-effects' models. For example, HB results can be very similar to 'model-averaging' estimates. HB can also trivially model individual heterogeneity in detection probabilities (the bane of most mark-recapture studies).
Many readers will be coming to this tutorial with some experience with Program MARK, which hails from the 'Frequentist' and IT-based 'model-averaging' school of analyses. In this tutorial we use the subjective Bayesian paradigm. The key distinction is that Mark provides fixed point-estimates of PCRD variables (like survival) which jointly "maximize the likelihood of having seen our data", and then does some brilliant Frequentist math to estimate 95%CIs for these variables. The Bayesian paradigm instead tries to estimate the probability distribution of our model variables, given that we saw the data that we saw. These probability distributions are called "Posterior Distributions", because they include information from our "Prior" that is updated with information from the data. You can do anything you like with a Posterior distribution: most people summarize it with a mean and 95% quantiles to approximate the point-estimates one encounters in Frequentism.
JAGS SYNTAX
Many of the same models in MARK can be run in JAGS. But whereas MARK employs 'PIMS' to parameterize and constrain models, the same thing is achieved more simply in the JAGS Bayesian Syntax (similar to R). For example, let's contrast two alternative parameterizations for a parameter 'phi' (survival), such as time-varying survival phi(t) versus time-invariant survival phi(dot). The two parameterizations can be represented in the following JAGS syntax (assuming with have 5 primary periods):
phi ~ dbeta(1,1) # time-invariant
for(t_ in 1:4){
phi_t[t_] <- phi
}
... contrasted with ...
for(t_ in 1:4){
phi_t[t_] ~ dbeta(1,1) # time-varying phi
}
What is going on? The first block of code declares that phi is a "random variable" (hence the tilde ~ operator) which has a prior distribution called the Beta distribution (all random variables need a prior distribution -- here, the Beta is useful for RV's that are probabilities, like the probability of surviving from one primary period to the other). There is only 1 phi, and we loop through 4 primary-period intervals (for(t_ in 1:4){ ...} ), during which phi is deterministically loaded (<-) into a vector called phi_t (hence, the <- operator). Each of the four elements of phi_i represent the survival between the 5 primary periods. Here, they are all equal(phi), hence 'time-invariant survival'.
In the second block of code, notice that we also have a vector called phi_t and a for loop, but instead of deterministically (<-) loading it with a single value (phi), each element of phi_t is itself an independent RV. Each has its own prior distribution, which all happen to be independent Beta distributions. Therefore, survival is allowed to be different between different primary periods. Simple!
PRIORS
Lets take a closer look at the Beta distribution and what the "dbeta(1,1)" means. The two 1's represent the shape parameters of the Beta distribution which controls the nature of our prior information. dbeta(1,1) looks flat on the probability scale, and is generally thought of as a default "vague" or "weak" prior for RV's like survival. It says our prior expectation is that E[phi]=0.5, but we aren't very confident. In fact, it is equivalent to only 1+1=2 'pseudo-observations': we are about as sure about phi as having just seen two obsevations of {alive,dead} (which isn't a lot of information). Alternatively, if we were more certain of a particular value, e.g., phi=0.8, we could use the (relatively) weak dbeta(4,1): now our prior expectation is E[phi]=0.8, and there is more probability density at high values, with a mode around 1. And yet, even dbeta(4,1) only amounts to about 4+1 'pseudo-observations' or {alive,alive,alive,alive,dead}. Our Posterior distribution will be a mixture of the evidence in the data (what we actually observed) and our prior information. Users must understand two key points: the less information we inject into our priors, the more our posterior point-estimates will be similar to the MLEs; and similarly the more information in our data (larger sample size, high detection probability), the more our posterior point-estimates will be similar to the MLEs. This has one very important consequences for PCRD users, and a key different between MARK and JAGS
- having a low detection probability means that you have very little information in your data; therefore, your posterior densities will very similar to your priors (so think carefully about them!).
- having a low detection probability means that you have very little information in your data; therefore, MARK often spits out stupid MLE estimates at boundary values (e.g., p = 1).
This is the key benefit of Bayesian Mark-Recapture under low-sample sizes and low-detection probabilities: whereas MARK often spits out garbage, the Bayesian posteriors will just return something similar to our prior information. What is the value of that?
WHO'S AFRAID OF MLE?
The problem with PCRD and Mark-recapture models for Maximum likelihood estimation is that these models are very parameter hungry, often 20 -30 parameters for a simple fixed-effect model. One is often in a situation of sparse data, low-sample sizes, and over-parametrization. Symptoms include boundary-level estimates, such as MLEs at 100% survival or 100% detectability, with meaningless estimates (Rankin et al. 2016). A 100% survival estimate is just plain stupid, despite being the Maximum Likelihood. Subjective Bayesian models (i.e., not using 'reference priors') help smooth over these boundary-level estimates. e.g., a MLE of p=1 will be pulled towards the prior expectation. Therefore, Bayesian models yield exact inference under any sample size. Of course, here is the catch: the prior plays a larger role under low-evidence/low-sample size situations.
But there is evidence from the Machine Learning community and those who care about prediction, that such weakly informative priors yeild better long-term predictions than MLEs alone. This is the problem with Frequentist methods: we are pretending our experiments are done in a vacuum, alone in the universe, without insight from long-term study and predictive assments. In this way, Subjective Bayesian models share something in common with "Regularization" employed in the Machine Learning community, a view that is explored in Hooten and Hobbs 2015.
BAYESIAN RESOURCES
Here is an accessible presentation Intro to Bayesian Inference and JAGS (Murdoch University R User Group, 2015). Marc Kery's WinBUGS for Ecologists is perhaps the best applied Bayesian books for non-statisticians, including R and BUGS code. Cressie et al (2009) and Halstead et al (2012) are two great articles motivating Bayesian inference for ecologists. Schofield and Barker (2011) and Schofield et al (2009) are accessible articles on mark-recapture generally in a Bayesian context. Michael Jordon has a great video on "Are You are Frequentist or Bayesian?".
This demo assumes the user has both R and JAGS installed, as well as the R package "rjags" to communicate between the two. See http:https://cran.r-project.org/ and http:https://mcmc-jags.sourceforge.net/. Most Linux distros have R/JAGS binaries available directly in standard repos. For example, Ubuntu users can simply type:
sudo apt-get update
sudo apt-get install r-base-core r-base-dev jags
Once in R, type:
install.package("rjags")
library("rjags")
source("R_PCRD_JAGS_SOURCE.R")
WARNING: Windows users should use navigate to the directory called WINDOWS to download the following tutorial files. Mac and GNU/Linux users can use the files in the root github directory.
There are 3 tutorials in 3 R files. Each tutorial presents a slightly different Bayesian model:
- R_PCRD_JAGS_firstcapt_fixedeff.R -- 'fixed-effect model with temporary migration (conditions on first capture)'
- R_PCRD_JAGS_fullcapt_fixedeff.R -- like above, but adds recruitment processes and full-capture modelling.
- R_PCRD_JAGS_hierarchical.R -- USERS SHOULD START HERE! A Hierarchical model that includes individual-heterogeneity and uses hyperpriors to achieve model parsimony (see the animation below)
The first file R_PCRD_JAGS_firstcapt_fixedeff.R is a tutorial for Pollock's Closed Robust Design, conditioning on first capture and is a "fixed-effects" version. The 2nd file R_PCRD_JAGS_fullcapt_fixedeff.R is also a fixed-effects version, but models the entire capture-history, thereby facilitating inference about recruitment processes (births). The 3rd file R_PCRD_JAGS_hierarchical.R is a Hierarchical Bayesian version, which will be of interest to most users: it can accommodate individual heterogeneity, full-capture modelling, and employs a peculiar type of "hyperprior" to induce a desirable shrinkage between fully time-varying parameters and time-constant parameters. In the companion paper, the authors argue that this hyperprior specification results in a type of "model-averaging" somewhat similar to the AICc-averaging that is ubiquitous in Mark-Recapture studies (thanks to the popularity of Program MARK).
Other files are:
- mark_capture_histories.inp -- real bottlenose dolphin data from Useless Loop, Western Australia
- R_PCRD_JAGS_SOURCE.R -- handy R code to initialize JAGS
- various ".JAG" files (model syntax)
The .inp file is a MARK data file containing capture histories for 5 years of photo-ID studies of bottlenose dolphins in the western gulf of Shark Bay, as presented originally in Nicholson et al. (2012). Please cite Nicholson when using this data. The SOURCE file contains some handy auxiliary functions to make it easier to initialize the JAGS models (especially generating sensible initial values for the latent-states of the HMM). The .JAG files are automatically generated when a user runs the above R tutorial scripts, and are not strictly necessary; they have the raw JAGS syntax for each JAGS model.
After cloning the appropriate files, users can re-analyze the data from Rankin et al. (2016) and Nicholson et al. (2012) by simply stepping through the R tutorials. Users should tweak the priors and model assumptions by customizing the JAGS model syntax (see the R files).
Users interested in either a) individual heterogeneity, or b) estimating recruitment processes should use the full-capture models. An advantage of the first-capture model is that users can model initial capture probabilities as being different from subsequent recapture probabilities (i.e., the p!=c specification in Program MARK). WARNING: users interested in the first-capture conditioning should familiarize themselves with the WinBUGS 'zeros trick'for exotic Likelihood functions (see http:https://users.aims.ac.za/~mackay/BUGS/Manuals/Tricks.html). Our First-capture models use a conditional multinomial distribution, the evaluation of which makes the JAGS Syntax somewhat more daunting for new users.
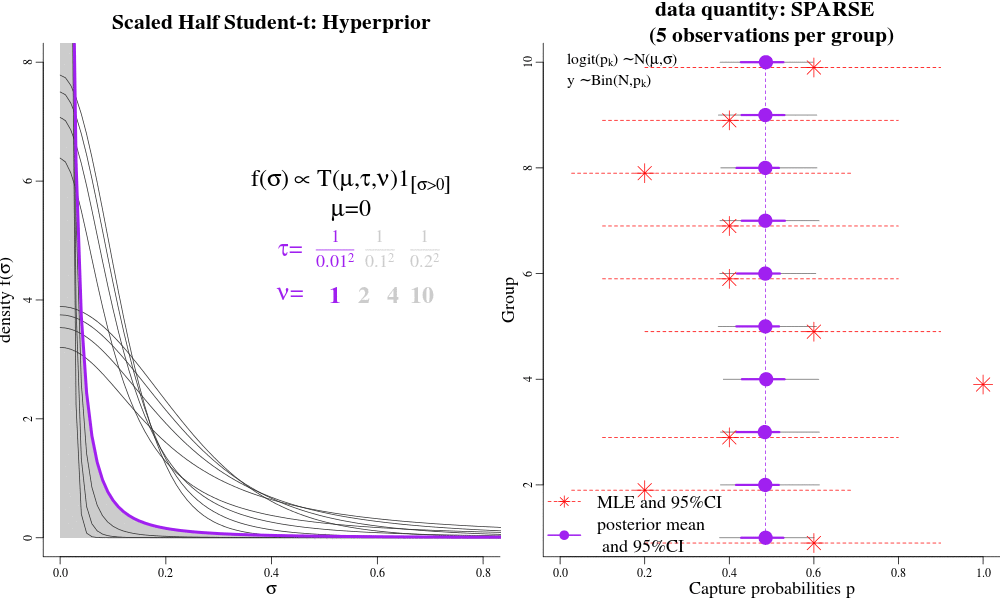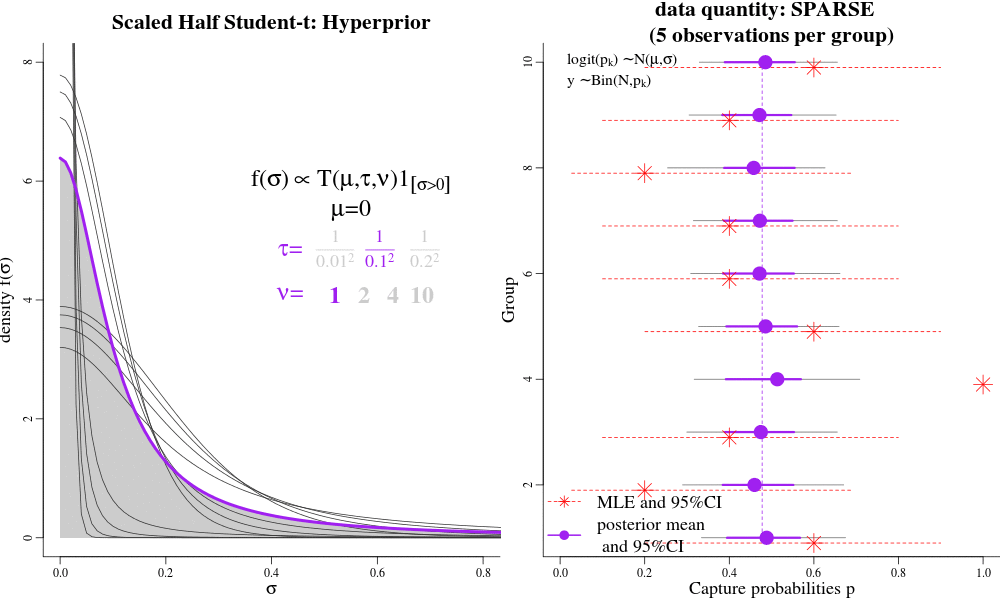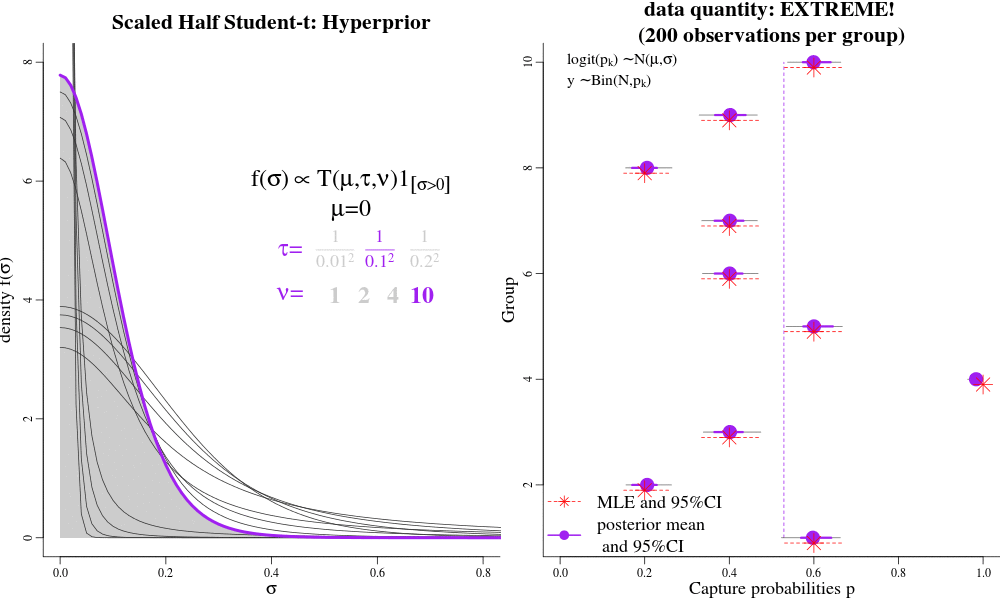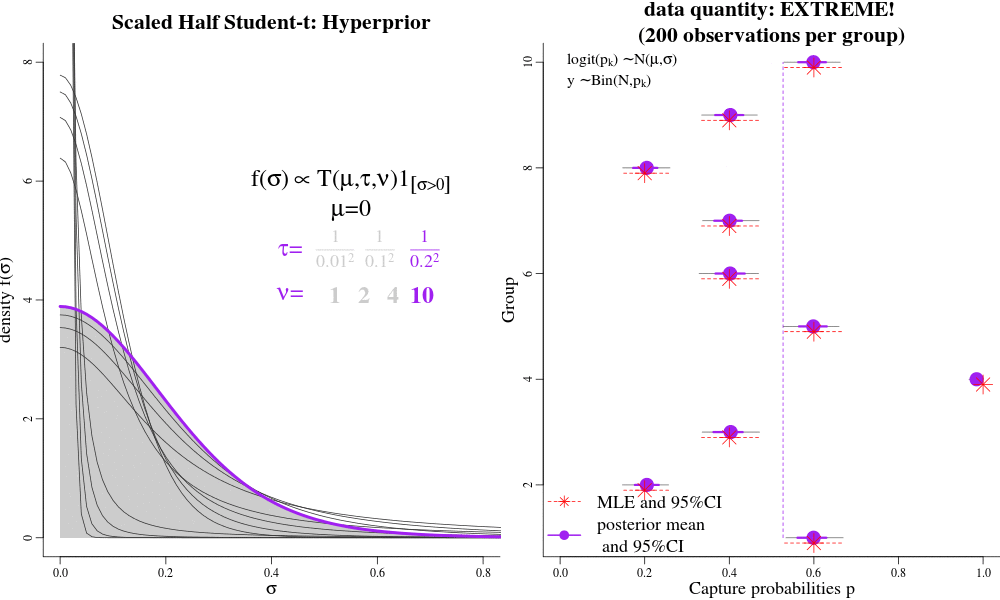
Users interested in the Hierarchical Bayesian (HB) model will likely want to change the "hyperparameters" that govern the amount of shrinkage between time-varying and time-constant parametrizations. The hyperparameters are passed to jags as 'data', called "pr.tauphi","pr.taug1","pr.taug2","pr.taupdmu","pr.taupd2nd","pr.taueps" in the JAGS syntax. Each of these hyperparameters govern a hyperprior distribution called the Scaled Half Student-T, with hyperparameters "mu","tau", and "nu". The half-T has favourable shrinkage-inducing properties, in that when there is little evidence in the data for widely-varying parameters, it shrinks a dispersion parameter to its prior expectation (near 0) and enforces a near time-constant parametrization for a particular parameter; conversely, lots of evidence in the data can result in large dispersion parameters and fully-time varying parameterizations, close to MLEs. Intermediate values for the Student T hyperparameters seem to result in parameter estimates similar to AICc model-averaged estimates.
See the animation. On the LEFT, there are different hyperpriors for sigma, each specified with different hyparameters. On the RIGHT, are the consequences of each hyperprior on individual p estimates. Under low-sample sizes, all the p's are "shrunk" to their overall group mean around 0.5. With more data, the p's approach their MLEs. This is parsimony: simple models under sparse data, and more complex models with larger sample sizes. This is often the goal of "model-averaging", but NOT for frequentist ML-based estimation.
For example in JAGS, the "pr.tauphi" hyperparameters will govern the disribution on "sigma.phi": if "sigma.phi" is close to zero, the survival "phi" will be nearly constant over time, and shrunk to its mean "phi.mu". Conversely, when "sigma.phi" is not zero, then "phi" will be fully time-varying, with individual phi values deviating strongly from their mean "phi.mu", and perhaps close to their individual Maximum Likelihood Estimates. The amount of shrinkage is controlled partially by the i) sample-size/amount of data, and ii) by the hyperparameters pr.tauphi=c(mean=0,tau=1/0.3^2,nu=2). Large values of 'tau' set the (approximate) prior expectation of 'sigma.phi', while 'nu' controls the tail of the distributions. Small values of 'nu' (2-3) allow the data more lee-way in deciding the value of 'sigma.phi'; large values of 'nu' make the hyper-prior more informative. A special case to avoid is nu=1, which is the half-Cauchy distribution, and allows too much dispersion for a random-variable that is between transformed between the logit and probability scales.
We recommend tau >= 0.3^-2 and nu => 2 as good starting points for the half Student-t hyper-parameters (especially detection probabilities and gamma-double-prime). Such values will result in parameter values that are close to the MLEs (if there is a lot data), but will still be slighly shrunk towards their time-constant values under weak/sparse data. However, for those parameters that are more difficult to identify and estimate, even with a lot of data, like survival and gamma-prime, it may be desireable to enforce more shrinkage. For example, in the bottlenose dolphin example, we had a prior expectation that there would be little year-to-year variation in apparent survival (survival should only vary 1-2% per year), and so we effectively coerced a near time-constant phi with the hyperparameters tau = 1/0.2^2, nu=13.
The above are hyperpriors, and therefore should be motivated by one's prior knowledge. Notice that in the hierarchical model, we place informative hyperpriors on the dispersion parameters, but admit little prior knowledge about the actual means of parameters.
Please don't run the models without first importing "R_PCRD_JAGS_SOURCE.R" into your R session.
source("R_PCRD_JAGS_SOURCE.R")
The file has a handy funtion to initialize JAGS variables. The Capture-Recapture models are possible in JAGS because we can re-parameterize the Capture-Recapture process as a state-space model (or Hidden Markov Model). This means we have a stochastic time-series of 'latent states' z={dead, inside, outside}. These are random variables just like any other random variable in the model. Unfortunately, it means that we must provide initial values of z for JAGS, in order to start the MCMC chains. This is a pain in the posterior for the novice user. Therefore, we have provided a handy forwards-messaging/backwards-sampling algorithm to estimate initial values of these latent states z.
If you modify or draw inspiration from our code, please cite JAGS, R, and the companion paper Rankin et al. (2016). If you use the bottlenose dolphin data from Useless Loop, please cite Nicholson et al. (2012). You can import the following into your reference manager:
@article{rankin_full-capture_2016, title = {A full-capture {Hierarchical} {Bayesian} model of {Pollock}'s {Closed} {Robust} {Design} and application to dolphins}, journal = {Frontiers in Marine Science}, author = {Rankin, Robert W. and Nicholson, Krista E. and Allen, Simon J. and Krützen, Michael and Bejder, Lars and Pollock, Kenneth H.}, year = {2016}, volume = {3}, number = {25}, url = {http:https://journal.frontiersin.org/article/10.3389/fmars.2016.00025}, doi = {10.3389/fmars.2016.00025} }
@article{nicholson_abundance_2012, title = {Abundance, survival and temporary emigration of bottlenose dolphins (\textit{{Tursiops} sp.}) off {Useless} {Loop} in the western gulf of {Shark} {Bay}, {Western} {Australia}}, volume = {63}, doi = {10.1071/MF12210}, number = {11}, journal = {Marine and Freshwater Research}, author = {Nicholson, Krista and Bejder, Lars and Allen, Simon J. and Krützen, Michael and Pollock, Kenneth H.}, year = {2012}, pages = {1059--1068} }
This project is possible because of the Shark Bay Dolphin Innovation Project. See http:https://www.sharkbaydolphins.org/?page_id=90.