- Questions
- Warm Up
- Load in Data
- Conduct Hypothesis Testing
YWBAT
- conduct a 1 samp and 2 samp ttest using scipy.stats
- test for criteria of 1 sample and 2 sample ttests
In a zoom chat, send an example of a type II error
- A probability that a data point would fall in to a set ASSUMING H0 IS TRUE.
- The probability of an event occurring given the null hypothesis is true.
- Pvalues change based on the problem
- There is a debate on the validity of using Pvalues...
import pandas as pd
import numpy as np
import scipy.stats as scs
from sklearn.datasets import load_iris
import statsmodels.api as sm
import statsmodels.stats as stats
import matplotlib.pyplot as plt
import seaborn as snsiris = load_iris()
data = iris.data
target = iris.target
features = iris.feature_namesdf = pd.DataFrame(data, columns=features)
df['target'] = target
df.head()
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
</style>
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | target | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0 |
df['target_names'] = df['target'].apply(lambda x: iris.target_names[x])df.head()
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
</style>
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | target | target_names | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0 | setosa |
df.target_names.unique()array(['setosa', 'versicolor', 'virginica'], dtype=object)
print(iris.DESCR).. _iris_dataset:
Iris plants dataset
--------------------
**Data Set Characteristics:**
:Number of Instances: 150 (50 in each of three classes)
:Number of Attributes: 4 numeric, predictive attributes and the class
:Attribute Information:
- sepal length in cm
- sepal width in cm
- petal length in cm
- petal width in cm
- class:
- Iris-Setosa
- Iris-Versicolour
- Iris-Virginica
:Summary Statistics:
============== ==== ==== ======= ===== ====================
Min Max Mean SD Class Correlation
============== ==== ==== ======= ===== ====================
sepal length: 4.3 7.9 5.84 0.83 0.7826
sepal width: 2.0 4.4 3.05 0.43 -0.4194
petal length: 1.0 6.9 3.76 1.76 0.9490 (high!)
petal width: 0.1 2.5 1.20 0.76 0.9565 (high!)
============== ==== ==== ======= ===== ====================
:Missing Attribute Values: None
:Class Distribution: 33.3% for each of 3 classes.
:Creator: R.A. Fisher
:Donor: Michael Marshall (MARSHALL%[email protected])
:Date: July, 1988
The famous Iris database, first used by Sir R.A. Fisher. The dataset is taken
from Fisher's paper. Note that it's the same as in R, but not as in the UCI
Machine Learning Repository, which has two wrong data points.
This is perhaps the best known database to be found in the
pattern recognition literature. Fisher's paper is a classic in the field and
is referenced frequently to this day. (See Duda & Hart, for example.) The
data set contains 3 classes of 50 instances each, where each class refers to a
type of iris plant. One class is linearly separable from the other 2; the
latter are NOT linearly separable from each other.
.. topic:: References
- Fisher, R.A. "The use of multiple measurements in taxonomic problems"
Annual Eugenics, 7, Part II, 179-188 (1936); also in "Contributions to
Mathematical Statistics" (John Wiley, NY, 1950).
- Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.
(Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.
- Dasarathy, B.V. (1980) "Nosing Around the Neighborhood: A New System
Structure and Classification Rule for Recognition in Partially Exposed
Environments". IEEE Transactions on Pattern Analysis and Machine
Intelligence, Vol. PAMI-2, No. 1, 67-71.
- Gates, G.W. (1972) "The Reduced Nearest Neighbor Rule". IEEE Transactions
on Information Theory, May 1972, 431-433.
- See also: 1988 MLC Proceedings, 54-64. Cheeseman et al"s AUTOCLASS II
conceptual clustering system finds 3 classes in the data.
- Many, many more ...
df.head()
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
</style>
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | target | target_names | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0 | setosa |
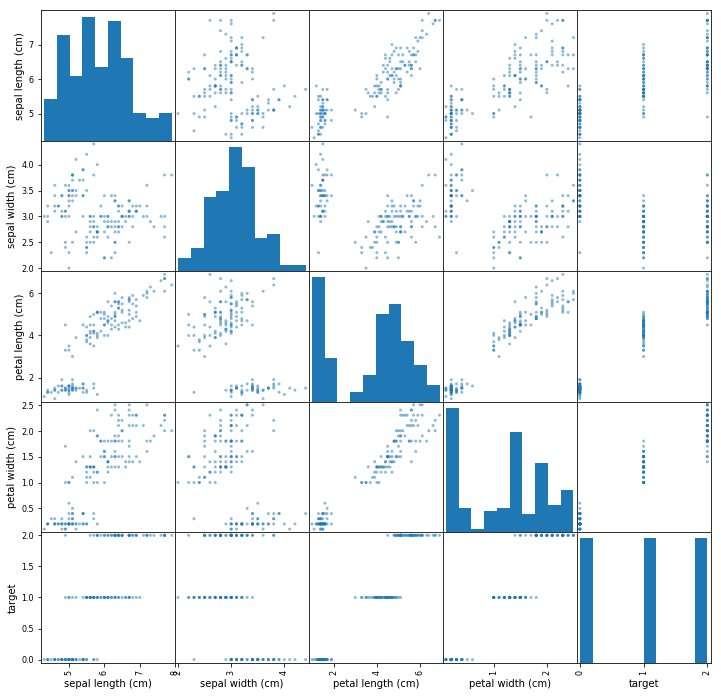
# pandas scattermatrix
pd.plotting.scatter_matrix(df, figsize=(12, 12))
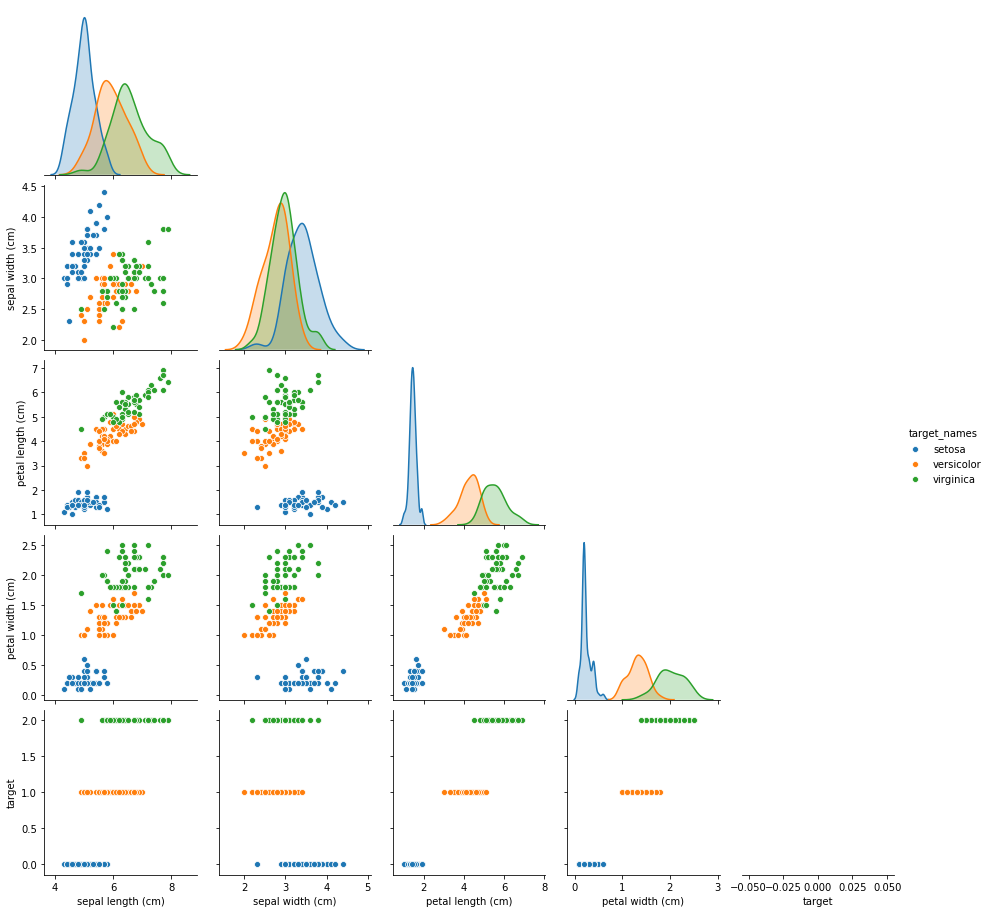
plt.show()sns.pairplot(df, hue='target_names', corner=True)
plt.show()/Users/rafael/anaconda3/envs/flatiron-env/lib/python3.6/site-packages/seaborn/distributions.py:288: UserWarning: Data must have variance to compute a kernel density estimate.
warnings.warn(msg, UserWarning)
- get data
- ensure it's normal
- take sampling statistics to compare the mean
- run levene and shapiro tests
- calculate cohen's d
- write up findings
sl_virg = df.loc[df['target_names']=='virginica', 'sepal length (cm)']
sl_vers = df.loc[df['target_names']=='versicolor', 'sepal length (cm)']- 2 sample ttest
- data is continuous - yes
- data follows normal distribution - we need to test
- variances are equal (otherwise you have to run a different ttest) - we need to test
- independent - yes
- sample things correctly and randomly - yes
-
H0:
$\mu_\text{virginica} = \mu_\text{versicolor}$ -
HA:
$\mu_\text{virginica} \neq \mu_\text{versicolor}$ -
H0: mu_virginica = mu_versicolor
-
HA: mu_virginica != mu_versicolor
sl_virg.shape, sl_vers.shape((50,), (50,))
# if i'm testing the means, I don't care about the populations
# what should I be taking
sample_mus_virginica = []
sample_mus_versicolor = []
for i in range(30):
virgSamp = np.random.choice(sl_virg, size=50, replace=True).mean()
versSamp = np.random.choice(sl_vers, size=50, replace=True).mean()
sample_mus_virginica.append(virgSamp)
sample_mus_versicolor.append(versSamp)# test each sample for normality using shapiro
# h0: data was drawn from a normal distribution
# ha: data was not drawn from a normal distribution
# if p<0.05 reject the null otherwise
# p >= 0.05 fail to reject the null
t1, p1 = scs.shapiro(sample_mus_virginica)
t2, p2 = scs.shapiro(sample_mus_versicolor)
print(p1)
print(p2)
# since p > 0.05 in both cases we fail to reject that the data is normal
# therefore we can assume that our data is normal0.9824318289756775
0.935992419719696
# test for equal variances using a levene test
# h0: samples are from populations with equal variances
# ha: samples are not from populations with equal variances
t, p = scs.levene(sample_mus_virginica, sample_mus_versicolor)
p
# since p>0.05 we fail to reject that the variances are equal
# therefore we can assume that our variances are equal0.33812261694131196
# Now that our assumptions are met we can actually do a ttest# h0: mu_virginica = mu_versicolor
# ha: mu_virginica != mu_versicolor
t, p = scs.ttest_ind(sample_mus_virginica, sample_mus_versicolor, equal_var=False)
p1.7437233880790833e-39
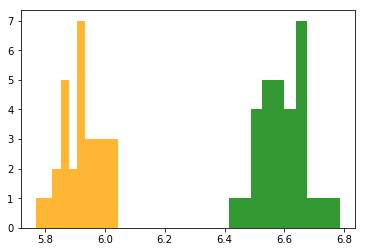
After doing a sampling distribution of the mean and running a welch's ttest one can conclude that the mean sample length of each group is different.
# how do we measure difference of means?
# power analysisdef cohen_d(x,y):
nx = len(x)
ny = len(y)
dof = nx + ny - 2
return np.abs(np.mean(x) - np.mean(y)) / np.sqrt(((nx-1)*np.std(x, ddof=1) ** 2 + (ny-1)*np.std(y, ddof=1) ** 2) / dof)effect_size = cohen_d(sample_mus_virginica, sample_mus_versicolor)
effect_size8.899510337442127
from statsmodels.stats.power import TTestIndPoweranalysis = TTestIndPower()result = analysis.solve_power(effect_size=effect_size, nobs1=30, alpha=0.05)
result1.0
type2error_rate = 1 - result
type2error_rate0.0
if you take a sample of versicolors or virginicas and measure the mean sepal length I can tell you with 100% certainity where the flowers came from.
plt.hist(sample_mus_virginica, color='green', alpha=0.8)
plt.hist(sample_mus_versicolor, color='orange', alpha=0.8)
plt.show()- don't only eyeball normality
- shapiro and levene test
- pairplots are awesome and very useful for data viz
- power analysis
- you'll always have 3 of 4 and it will always solve for the 4th
- input into the scatter matrix
- cohen's d to measure the effect size
- alpha=0.05 because it's standard
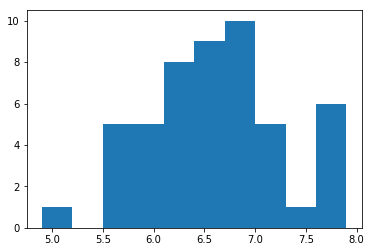
plt.hist(sl_virg)
plt.show()sl_virg_zscores = scs.zscore(sl_virg)
random_zscores = np.random.normal(0, 1, 50)# H0: sl_virg = normal distribution
# HA: sl_virg != normal distribution
scs.kstest(sl_virg_zscores, 'norm', args=(0, 1))
# pvalue of 0, which means we reject the nullKstestResult(statistic=0.11558941328638006, pvalue=0.48764984405744605)
random_normal_data = np.random.normal(50, 10, 1000)# H0: random_normal_data = normal distribution
# HA: random_normal_data != normal distribution
scs.kstest(random_normal_data, 'norm', args=(50, 10))
# pvalue of 0, which means we reject the nullKstestResult(statistic=0.03540056710981665, pvalue=0.15929441756654197)
def ks_test(arr):
zscores_arr = scs.zscore(arr)
t, p = scs.kstest(zscores_arr, 'norm', args=(0, 1))
return t, pks_test(sl_virg)(0.11558941328638006, 0.48764984405744605)
scs.anderson(sl_virg, 'norm')AndersonResult(statistic=0.5516407106574732, critical_values=array([0.538, 0.613, 0.736, 0.858, 1.021]), significance_level=array([15. , 10. , 5. , 2.5, 1. ]))
Shapiro Wilks is less strict than a KS Test
- use both
When
- Multiple Comparisons
- Number of Ice Cream sold by Flavors
- Sepal Length of all iris flowers (3)
- Drug Testing (more than 2 groups)
- Testing more than 2 groups
What does it tell you?
- H0: u1 = u2 = ... = uN
- HA: ui!=uj for some i and j in our groups
sl_seto = df.loc[df['target_names']=='setosa', 'sepal length (cm)']sl_sd_virg = []
sl_sd_vers = []
sl_sd_seto = []
for i in range(30):
s1 = np.random.choice(sl_seto, 50, replace=True).mean()
s2 = np.random.choice(sl_vers, 50, replace=True).mean()
s3 = np.random.choice(sl_virg, 50, replace=True).mean()
sl_sd_seto.append(s1)
sl_sd_vers.append(s2)
sl_sd_virg.append(s3)for samp in [sl_sd_seto, sl_sd_vers, sl_sd_virg]:
t, p = ks_test(samp)
print(t, p)
# pvalues > 0.05 -> data is normal0.07106849093676221 0.9981258672951128
0.10135268270444081 0.9175703455910869
0.1179278228199826 0.7983071102773645
# H0: input samples are from equal variances
# HA: input samples are not from equal variances
scs.bartlett(sl_sd_seto, sl_sd_vers, sl_sd_virg)
# p<=0.05 -> reject the null variances are not equal the differenc of spreads isn't greater than 3BartlettResult(statistic=17.335422788020647, pvalue=0.00017205241417272234)
# H0: u1 = ... = un
# HA: ui!=uj
scs.f_oneway(sl_sd_seto, sl_sd_vers, sl_sd_virg)
# p<0.05 reject the nullF_onewayResult(statistic=3518.8142885838843, pvalue=5.941971274788364e-84)
df['sl'] = df['sepal length (cm)']import statsmodels.api as sm
from statsmodels.formula.api import olsmodel = ols('sl ~ C(target_names)', data=df).fit()
anova_table = sm.stats.anova_lm(model, typ=2)
anova_table
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
</style>
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| sum_sq | df | F | PR(>F) | |
|---|---|---|---|---|
| C(target_names) | 63.212133 | 2.0 | 119.264502 | 1.669669e-31 |
| Residual | 38.956200 | 147.0 | NaN | NaN |
the sepal of at least one group is different than the another group
mc = stats.multicomp.MultiComparison(df['sepal length (cm)'], df['target_names'])res = mc.tukeyhsd()
print(res) Multiple Comparison of Means - Tukey HSD, FWER=0.05
=========================================================
group1 group2 meandiff p-adj lower upper reject
---------------------------------------------------------
setosa versicolor 0.93 0.001 0.6862 1.1738 True
setosa virginica 1.582 0.001 1.3382 1.8258 True
versicolor virginica 0.652 0.001 0.4082 0.8958 True
---------------------------------------------------------
for col in df.columns[:4]:
mc = stats.multicomp.MultiComparison(df[col], df['target_names'])
print(f"Testing: {col}")
res = mc.tukeyhsd()
print(res)
print("\n\n")Testing: sepal length (cm)
Multiple Comparison of Means - Tukey HSD, FWER=0.05
=========================================================
group1 group2 meandiff p-adj lower upper reject
---------------------------------------------------------
setosa versicolor 0.93 0.001 0.6862 1.1738 True
setosa virginica 1.582 0.001 1.3382 1.8258 True
versicolor virginica 0.652 0.001 0.4082 0.8958 True
---------------------------------------------------------
Testing: sepal width (cm)
Multiple Comparison of Means - Tukey HSD, FWER=0.05
============================================================
group1 group2 meandiff p-adj lower upper reject
------------------------------------------------------------
setosa versicolor -0.658 0.001 -0.8189 -0.4971 True
setosa virginica -0.454 0.001 -0.6149 -0.2931 True
versicolor virginica 0.204 0.0088 0.0431 0.3649 True
------------------------------------------------------------
Testing: petal length (cm)
Multiple Comparison of Means - Tukey HSD, FWER=0.05
=========================================================
group1 group2 meandiff p-adj lower upper reject
---------------------------------------------------------
setosa versicolor 2.798 0.001 2.5942 3.0018 True
setosa virginica 4.09 0.001 3.8862 4.2938 True
versicolor virginica 1.292 0.001 1.0882 1.4958 True
---------------------------------------------------------
Testing: petal width (cm)
Multiple Comparison of Means - Tukey HSD, FWER=0.05
=========================================================
group1 group2 meandiff p-adj lower upper reject
---------------------------------------------------------
setosa versicolor 1.08 0.001 0.9831 1.1769 True
setosa virginica 1.78 0.001 1.6831 1.8769 True
versicolor virginica 0.7 0.001 0.6031 0.7969 True
---------------------------------------------------------