RiboDoc is a bioinformatics pipeline for Ribosome sequencing (Ribo-seq) data. It can be used to perform quality control, trimming, alignment and downstream qualitative and quantitative analysis.
It can be used with multiple operating systems, and it's goal is to standardize the general steps that must be performed systematically in Ribo-seq analysis, together with the statistical analysis and quality control of the sample. The data generated can then be exploited with more specific tools.
RiboDoc is a tool designed to standardize bioinformatics analyses in the field of translation, following the FAIR guidelines to make installation and analysis meet principles of findability, accessibility, interoperability, and reusability. Thus, this pipeline is built using Snakemake, a workflow management system to create reproducible and scalable data analyses. Additionally, it is a Docker-based package, which means it can be used by anyone. Docker is a container which packages up code and all its dependencies so RiboDoc can run quickly and reliably from one computing environment to another.
If you want to easily understand how to launch RiboDoc on your own computer, you can check our video tutorial just here :

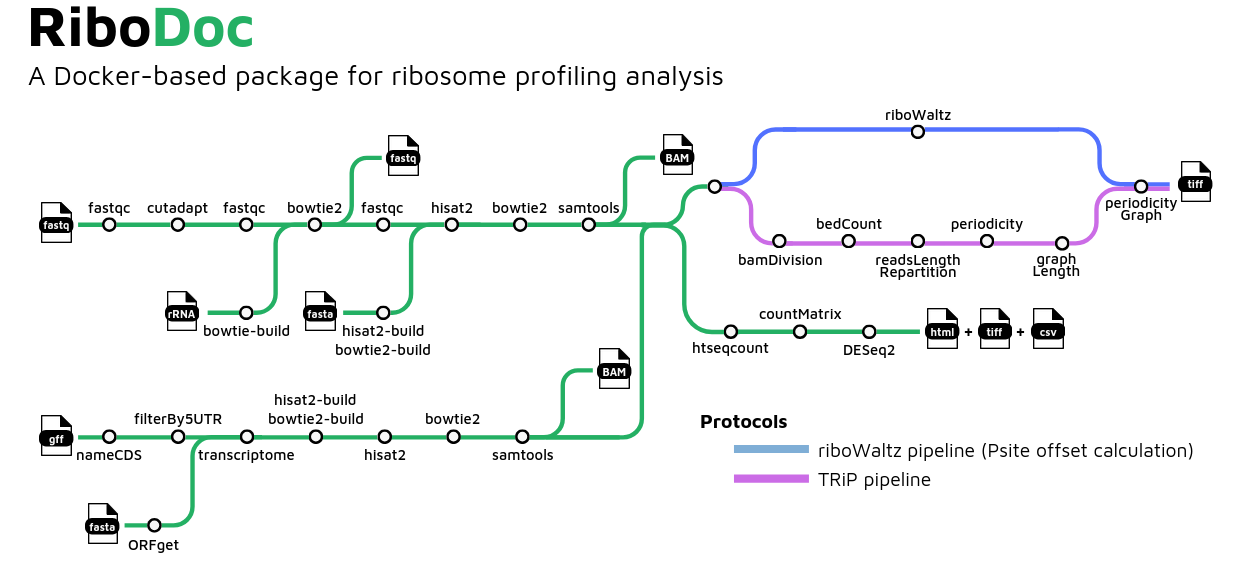
RiboDoc is designed to perform all classical steps of ribosome profiling (RiboSeq) data analysis from the FastQ files to the differential expression analysis with necessary quality controls.
- Quality Control of raw reads with
FastQC - Adapter and quality trimming, read length filtering with
Cutadapt - Quality Control of trimmed reads with
FastQC - Removal of contaminants RNA (rRNA, tRNA, viral RNA, ...) with
Bowtie2 - Quality Control of depleted reads with
FastQC - Genome and transcriptome alignment of reads conjointly with
Hisat2andBowtie2 - Sort and index alignments with
samtools - Reads Count with
htseq-count9.Analysis of differential gene expression with 'DESeq2' - Offset prediction and periodicity graph creation with
ribowaltzorTRiP
-
Ensure
DockerorSingularityare installed on your system. If you don't have super user rights (if your work on a cluster for example), Singularity might be prefered as it does not required it. -
A precise architecture in your project folder is required. The first step is the project folder creation. It is named as your project and will be the volume linked to the container. Then, two sub-folders and a file have to be created and filled.
Caution, those steps are majors for the good course of the analysis. The subfolders names do not have uppercase letters.
a. Create the first subfolder and name it fastq. This subfolder, as its name suggests, should contain your FastQ files compressed in gzip format (*.gz*).
Format of file names must be as following:
[CONDITION]_[NAME].[REPLICATE].fastq.gz
For example, a replicate of the wild-type condition the sample could be named Wild_Type.56.fastq.gz and the name of a replicate for the mutant samples could be Mutant.42.fastq.gz