Note: The COVID-Net models provided here are intended to be used as reference models that can be built upon and enhanced as new data becomes available. They are currently at a research stage and not yet intended as production-ready models (not meant for direct clinical diagnosis), and we are working continuously to improve them as new data becomes available. Please do not use COVID-Net for self-diagnosis and seek help from your local health authorities.
Recording to webinar on How we built COVID-Net in 7 days with Gensynth
Update 04/16/2020: If you have questions, please check the new FAQ page first.
Update 04/15/2020: We released two new models, COVIDNet-CXR Small and COVIDNet-CXR Large, which were trained on a new COVIDx Dataset with both PA and AP X-Rays from Cohen et al, as well as additional COVID-19 X-Ray images from Figure1.
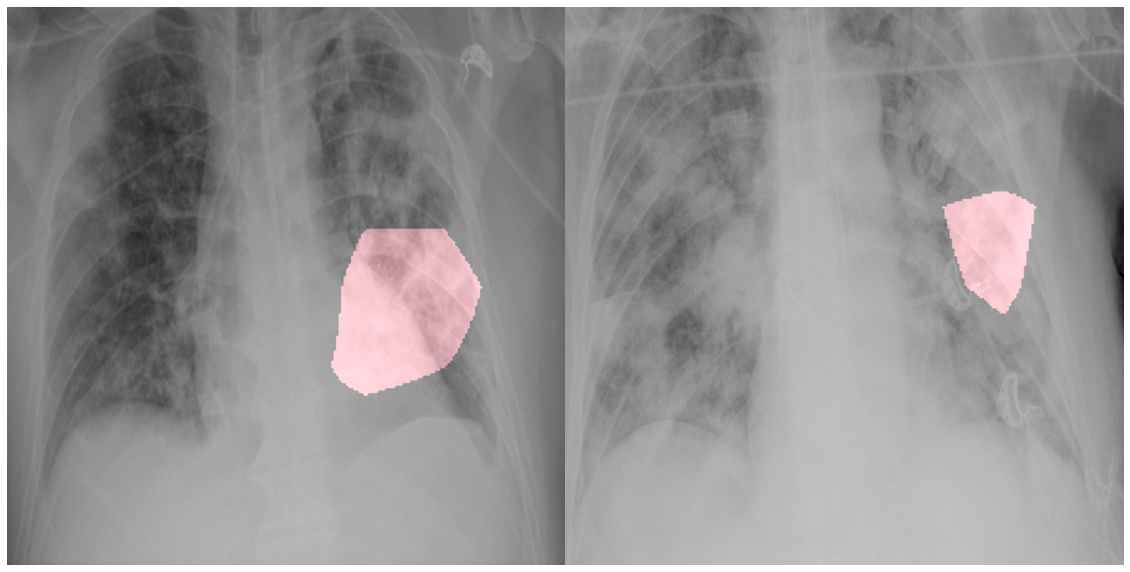
Example chest radiography images of COVID-19 cases from 2 different patients and their associated critical factors (highlighted in red) as identified by GSInquire.
The COVID-19 pandemic continues to have a devastating effect on the health and well-being of the global population. A critical step in the fight against COVID-19 is effective screening of infected patients, with one of the key screening approaches being radiological imaging using chest radiography. It was found in early studies that patients present abnormalities in chest radiography images that are characteristic of those infected with COVID-19. Motivated by this, a number of artificial intelligence (AI) systems based on deep learning have been proposed and results have been shown to be quite promising in terms of accuracy in detecting patients infected with COVID-19 using chest radiography images. However, to the best of the authors' knowledge, these developed AI systems have been closed source and unavailable to the research community for deeper understanding and extension, and unavailable for public access and use. Therefore, in this study we introduce COVID-Net, a deep convolutional neural network design tailored for the detection of COVID-19 cases from chest radiography images that is open source and available to the general public. We also describe the chest radiography dataset leveraged to train COVID-Net, which we will refer to as COVIDx and is comprised of 13,800 chest radiography images across 13,725 patient patient cases from three open access data repositories. Furthermore, we investigate how COVID-Net makes predictions using an explainability method in an attempt to gain deeper insights into critical factors associated with COVID cases, which can aid clinicians in improved screening. By no means a production-ready solution, the hope is that the open access COVID-Net, along with the description on constructing the open source COVIDx dataset, will be leveraged and build upon by both researchers and citizen data scientists alike to accelerate the development of highly accurate yet practical deep learning solutions for detecting COVID-19 cases and accelerate treatment of those who need it the most.
For a detailed description of the methodology behind COVID-Net and a full description of the COVIDx dataset, please click here.
Currently, the COVID-Net team is working on COVID-RiskNet, a deep neural network tailored for COVID-19 risk stratification. Currently this is available as a work-in-progress via included train_risknet.py script, help to contribute data and we can improve this tool.
If you would like to contribute COVID-19 x-ray images, please submit to https://figure1.typeform.com/to/lLrHwv. Lets all work together to stop the spread of COVID-19!
If you are a researcher or healthcare worker and you would like access to the GSInquire tool to use to interpret COVID-Net results on your data or existing data, please reach out to [email protected] or [email protected]
Our desire is to encourage broad adoption and contribution to this project. Accordingly this project has been licensed under the GNU Affero General Public License 3.0. Please see license file for terms. If you would like to discuss alternative licensing models, please reach out to us at [email protected] and [email protected] or [email protected]
If there are any technical questions after the README, FAQ, and past/current issues have been read, please post an issue or contact:
If you find our work useful, can cite our paper using:
@misc{wang2020covidnet,
title={COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images},
author={Linda Wang, Zhong Qiu Lin and Alexander Wong},
year={2020},
eprint={2003.09871},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
- DarwinAI Corp., Canada and Vision and Image Processing Research Group, University of Waterloo, Canada
- Linda Wang
- Alexander Wong
- Zhong Qiu Lin
- Paul McInnis
- Audrey Chung
- Hayden Gunraj, COVIDNet for CT: Coming soon.
- Vision and Image Processing Research Group, University of Waterloo, Canada
- James Lee
- Matt Ross and Blake VanBerlo (City of London), COVID-19 Chest X-Ray Model: https://github.com/aildnont/covid-cxr
- Ashkan Ebadi (National Research Council Canada)
- Kim-Ann Git (Selayang Hospital)
- Abdul Al-Haimi
- Requirements to install on your system
- How to generate COVIDx dataset
- Steps for training, evaluation and inference
- Results
- Links to pretrained models
The main requirements are listed below:
- Tested with Tensorflow 1.13 and 1.15
- OpenCV 4.2.0
- Python 3.6
- Numpy
- Scikit-Learn
- Matplotlib
Additional requirements to generate dataset:
- PyDicom
- Pandas
- Jupyter
These are the final results for COVIDNet-CXR Small and COVIDNet-CXR Large.

Confusion matrix for COVIDNet-CXR Small on the COVIDx test dataset.
| Sensitivity (%) | ||
|---|---|---|
| Normal | Pneumonia | COVID-19 |
| 97.0 | 90.0 | 87.1 |
| Positive Predictive Value (%) | ||
|---|---|---|
| Normal | Pneumonia | COVID-19 |
| 89.8 | 94.7 | 96.4 |

Confusion matrix for COVIDNet-CXR Large on the COVIDx test dataset.
| Sensitivity (%) | ||
|---|---|---|
| Normal | Pneumonia | COVID-19 |
| 99.0 | 89.0 | 96.8 |
| Positive Predictive Value (%) | ||
|---|---|---|
| Normal | Pneumonia | COVID-19 |
| 91.7 | 98.9 | 90.9 |