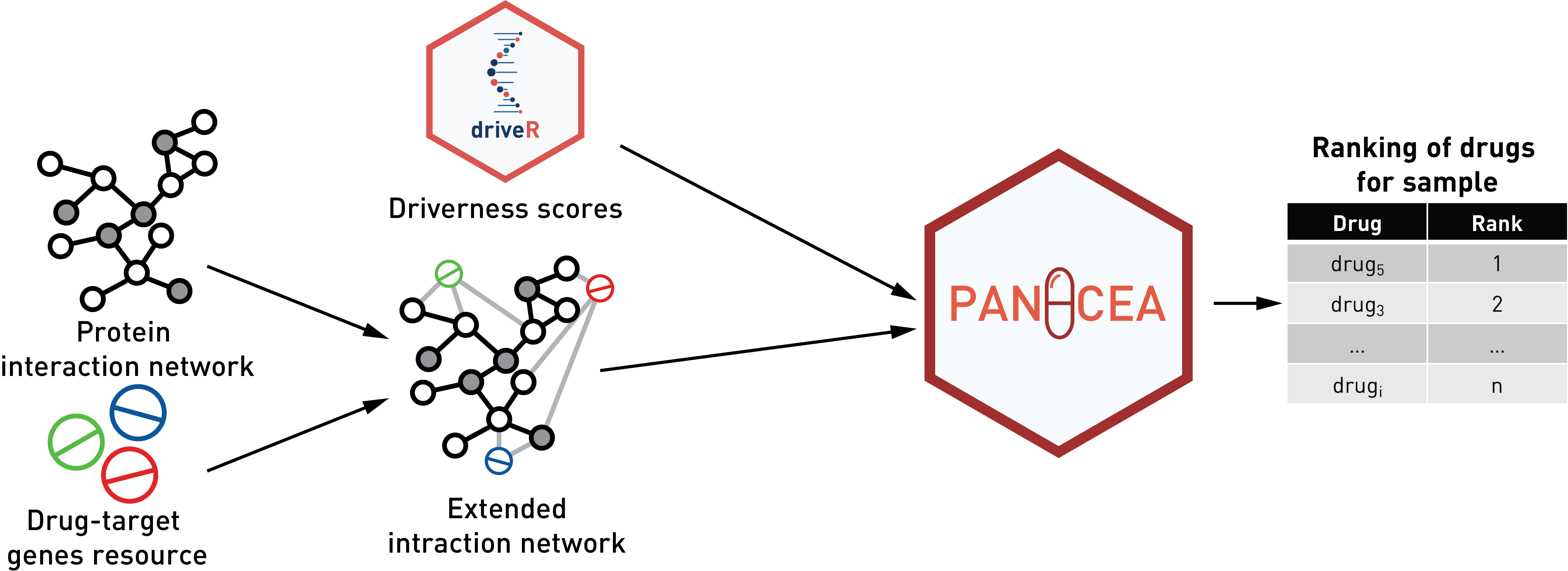
Identification of the most appropriate pharmacotherapy for each patient
based on genomic alterations is a major challenge in personalized
oncology. PANACEA is a collection of personalized anti-cancer drug
prioritization approaches utilizing network methods. The methods utilize
personalized “driverness” scores from
driveR to rank drugs, mapping
these onto a protein-protein interaction network. The “distance-based”
method scores each drug based on these scores and distances between
drugs and genes to rank given drugs. The “RWR” method propagates these
scores via a random-walk with restart framework to rank the drugs.
The method is described in detail in Ulgen E, Ozisik O, Sezerman OU. PANACEA: network-based methods for pharmacotherapy prioritization in personalized oncology. Bioinformatics. 2023 Jan 1;39(1):btad022. https://doi.org/10.1093/bioinformatics/btad022
PANACEA workflowYou can install the latest release version of PANACEA from CRAN via:
install.packages("PANACEA")You can install the development version of PANACEA from GitHub with:
# install.packages("devtools") @ if you don't have devtools installed
devtools::install_github("egeulgen/PANACEA", build_vignettes = TRUE)The wrapper function score_drugs() can be used to score and rank drugs
for an individual tumor sample via the “distance-based” or “RWR” method.
The required inputs are:
driveR_res: data frame of driveR results. Details on how to obtaindriveRoutput are provided in this vignettedrug_interactions_df: data frame of drug-gene interactions (defaults to interactions from DGIdb expert-curated sources)W_mat: (symmetric) adjacency matrix for the protein interaction network (defaults to STRING v11.5 interactions with combined score > .4)method: scoring method (one of “distance-based” or “RWR”)
For detailed information on how to use PANACEA, please see the
vignette “How to use PANACEA” via vignette("how_to_use") or visit
this link