This section is done from any terminal on cori, usually by doing something like:
- log into cori and follow instructions here
-
Go to your
$HOMEdirectory:cd $HOME -
Create a symlink to Forrest Hoffman's downloaded data:
ln -s /global/cscratch1/sd/cmip6 cmip6_data -
clone the git repo to get the examples and tutorials:
git clone git:https://github.com/doutriaux1/rgma_cdat_tutorial
I update the repo frequently to get the latest changes you will probably need to revert the changes generated locally by running the notebooks.
cd $HOME/rgma_cdat_tutorial
git checkout -- .
git pull
This section is done within a browser, preferably Chrome or Firefox.
- log to jupyter-hub at: https://jupyter.nersc.gov/hub/login?next=%2Fhub%2Fhome
You should see something like the picture below
- Find the "Jupyterlab" file navigator (folder icon at top left, bellow "orange Jupyter" icon and above "github cat icon")
- click on the "home"/"root" button of this navigator.
- then navigate back to your home folder via the
homedirectory, followed by thejovyandirectory.jovyanis the container's user but it is mounted onto (linked to) your NERSC$HOMEdirectory.
-
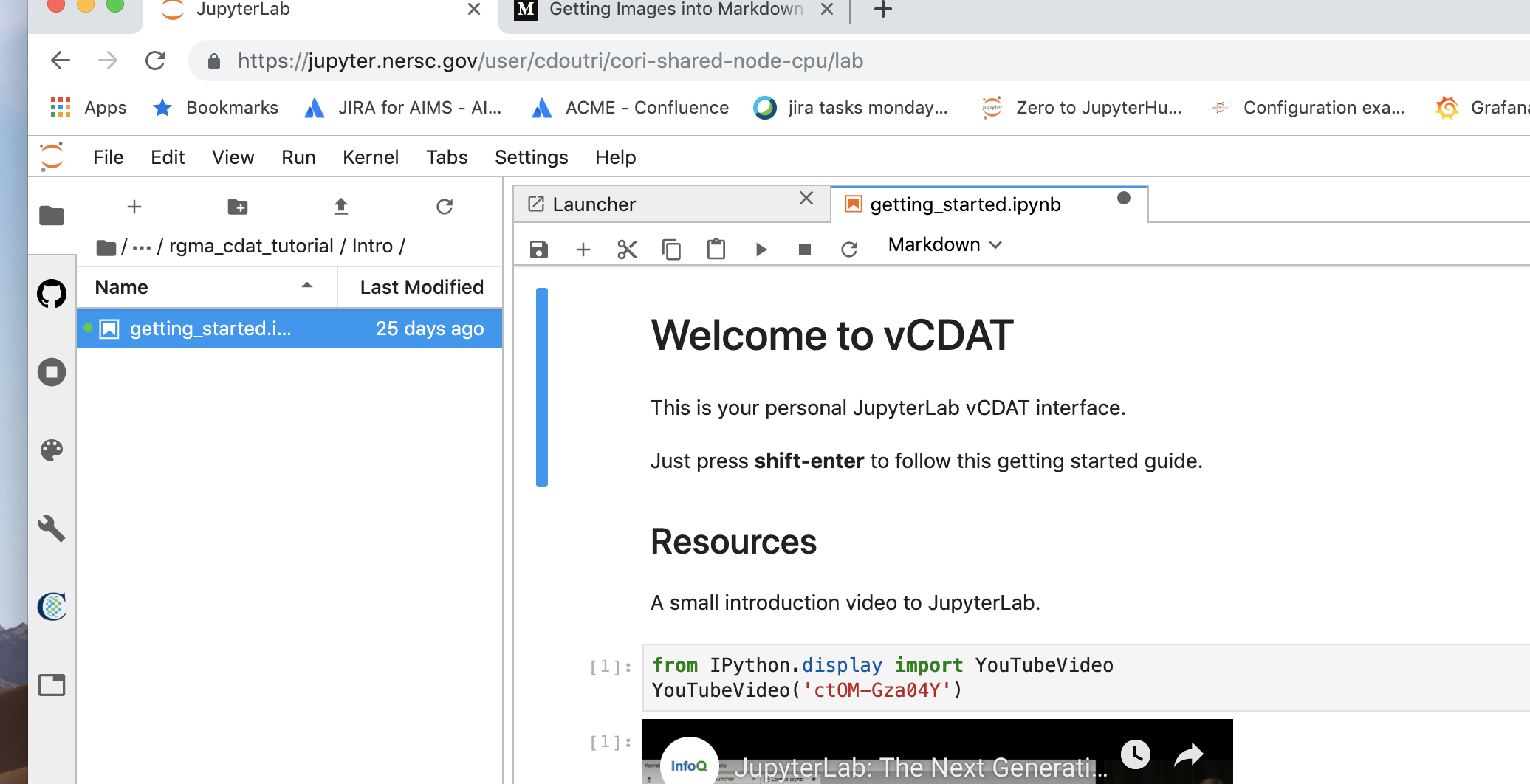
In the Jupyterlab navigator, go to home and look for the
rgma_cdat_tutorialfolder. This folder contains our notebooks for the CDAT tutorial. -
Navigate down to the
Introdirectory, and click on the getting started notebook. Feel free to watch the video before the class.