This is a repository containing files to register one IVUS pullback with a second IVUS pullback. Typically these pullbacks are taken at 2 different timepoints such as baseline (BL) and follow-up (FU). The code registers IVUS pullbacks in both the longitudinal and circumferential directions. In order to do this the code employs a 3D graph where an optimal path is found representing the optimum (with respect to the cost function) matching of images along the axis and along the circumferential direction. The methodology employed here is based on two papers and any use of this work should consider citing the following papers.
- Framework to co-register longitudinal virtual histology-intravascular ultrasound data in the circumferential direction. Timmins LH, Suever JD, Eshtehardi PE, McDaniel MC, Oshinski JN, Samady H, Giddens DP. 2013. IEEE Trans Med Img, Vol 32(11)
- Simultaneous Registration of Location and Orientation in Intravascular Ultrasound Pullback Pairs Via 3D Graph-Based Optimization. Zhang L, Wahle A, Chen Z, Zhang L, Downe RW, Kovarnik T, Sonka M. 2015. IEEE Trans Med Img, Vol 34(12)
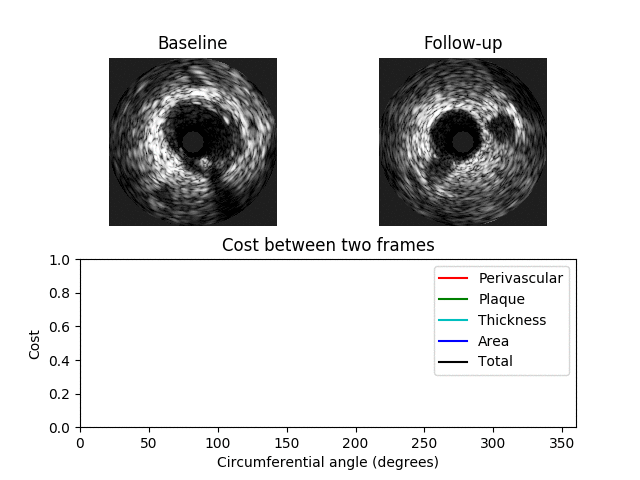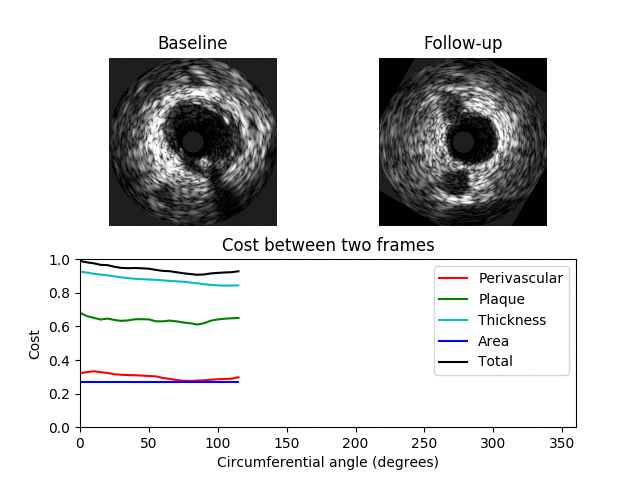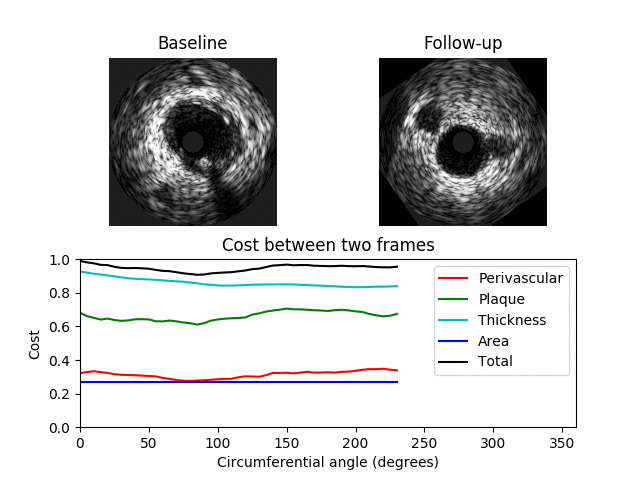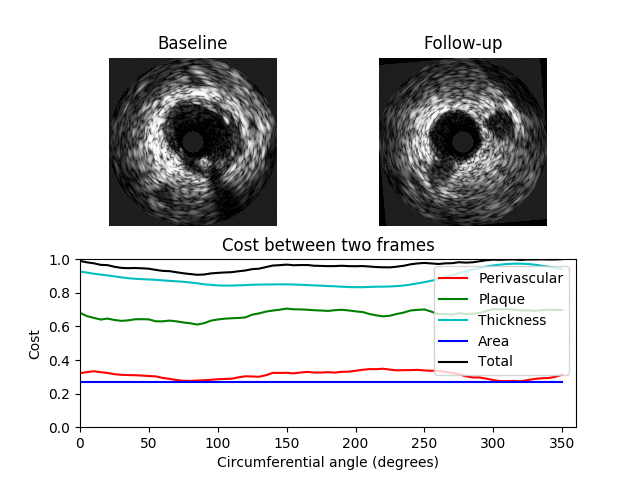
A 3D graph is constructed where a value in the graph represents the similarity between two IVUS frames at a specific orientation where a value of 0 indicates a perfect match. The 3D graph has 3 axes: x-axis -> BL image; y-axis -> angle; z-axis -> FU image. The value C[i, j, k] contains the cost between the ith BL image and the kth FU image when the FU image is rotated by j degrees The 3D graph/matrix C is generated from costs between image features at different orientations. The costs are computed for each feature separately and are weighted by user-specified values. A cumulative cost matrix CC is then determined through dynamic programming. The minimal cost path through the cumulative cost matrix corresponds to the optimum registration between the BL and FU IVUS pullbacks.
 Minimum cost path through cumulative cost matrix
Minimum cost path through cumulative cost matrix
Four features are used to register the pullbacks. These are:
- The perivascular tissue
- The plaque tissue
- The plaque thickness
- The lumen area
Examples showing when the cost is low (top) and high (bottom) between two different images are shown below. Here a minimum total cost of 0.4 is found when the FU image is rotated by 130 degrees. In the second example the minimum cost is 0.92 indicating a poor match between the images.
Each IVUS frame must be a .tif/jpg file and contours are required. Files must be named by their frame number beginning from 0 e.g. 10.jpg is the 11th frame of the pullback. Users should open registration.py and enter the patient/case name
patient_name = 'my_patient'Following this the user should enter the path to the IVUS images and contours
# BL files
BLpath = 'path/to/BL/tif'
BLxmlpath = 'path/to/BL/xml'
# FU files
FUpath = 'path/to/FU/tif'
FUxmlpath = 'path/to/FU/xml'Finally the user can enter the start and end frames automatic start and end frame detection is present however, the robustness of this is still under investigation.
# start and end frames in format [BL, FU]
start_idx = [5, 28]
end_idx = [122, 145]After successful completion each registered FU image is rotated to match the corresponding BL image orientation and saved to file. Finally, a file pname_registered.csv is saved to the current directory. The file has the following format
BL frame number, FU frame number, rotation angle
BL and FU images as well as their contours are provided. To run the demo from the downloaded directory run:
python registration.pyFormat used by Echoplaque. Stores meta-information and contour coordinates. If you have contours of IVUS images, each image needs to be converted into a label mask (see sample mask in docs). Each mask should have the same dimensions as an IVUS image. Pixels in the image corresponding to lumen, plaque, perivascular and background should have values corresponding to 1, 2, 3 and 0 respectively. To create an XML file for a pullback of 131 frames with a frame resolution of 0.02mm and acquired with a pullback speed of 0.5mm/s run the following
python write_xml.py --dir='path/to/masks' --res=0.02 --frames=131 --speed=0.5Requires python2
pip install -r requirements.txt