- Introduction
- Data description
- Notebooks
- 00_setup.ipynb
- 01_data_cleaning.ipynb
- 02_data_exploration.ipynb
- 03_data_augmentation.ipynb
- 04_baseline_model
- 05_metadata.ipynb
- 06_multilabel_classification.ipynb
- 07a_diff_archs_resnet101.ipynb
- 07b_diff_archs_densenet121.ipynb
- 07c_diff_archs_alexnet.ipynb
- 08_weight_decay_and_tta.ipynb
- 09_mixed_precision.ipynb
- 10_progressive_resizing.ipynb
- 11_model_deployment.ipynb
- Papers Read
- References
Hemorrhage in the brain (Intracranial Hemorrhage) is one of the top five fatal health problems. The hemorrhage causes bleeding inside the skull (typically known as cranium). It accounts for approximately 10% of strokes in the U.S thus diagnosing it quickly and efficiently is of utmost importance.
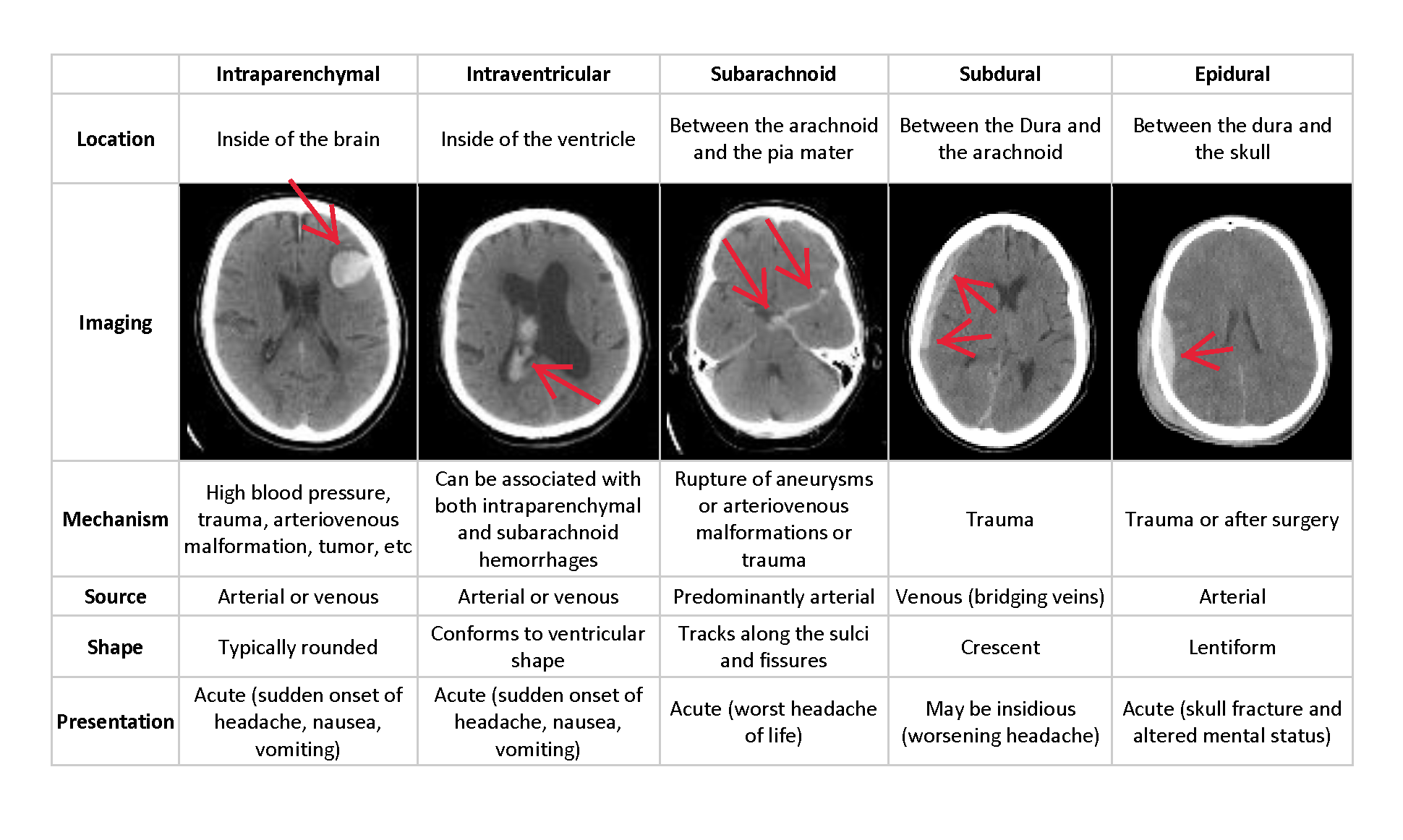
Intracranial Hemorrhage can have many consequences depending on the location, size, type of bleed in the brain. The relationship is not that simple to interpret. A small hemorrhage might be equally fatal as a big one if it is in a critical location. If a patient shows acute hemorrhage symptoms such as loss of consciousness then an immediate diagnosis is required to carry out the initial tests but the procedure is very complicated and often time-consuming. The delay in diagnosis and treatment can have severe effects on patients, especially the ones in need of immediate attention. The solution proposed in this project aims to make this process efficient by automating the preliminary step to identify the location, type, and size of the bleed, the doctors can provide a better diagnosis and a faster one as the work of identification is done by the algorithm. Also, the algorithm treats every scan with the same scrutiny, reducing human error and learning more as it goes.
Thus, the challenge is to build an algorithm to detect acute intracranial hemorrhage and its subtypes. This project will be a step towards identifying each of these relevant attributes for a better diagnosis.
Check the image below for more information on the various subtypes.
We use Jeremy Howard's clean dataset instead of the original dataset An in depth analysis of how he created this dataset is shown here. Jeremy's dataset has an image size of (256,256) and the metadata stored in data frames. For any troubles downloading the original dataset, refer to this link
The data is provided in 3 parts, the dicom images that look as follows
metadata such as Patient ID, Image position, Image Orientation, window center, window width and so on. And a target data frame mapping every patient to the hemorrhage.
For domain knowledge, refer to readiology masterclass
The kernels are arranged in numerical order so that people can see where we started and how we went about our experiments
00_setup.ipynb - how to setup and download the dataset on Google colab.
01_data_cleaning.ipynb - steps followed to clean the data
02_data_exploration.ipynb- exploratory data analysis on the images and csv files
03_data_augmentation.ipynb - applying various transforms on the images and seeing which ones are useful.
04_baseline_model - train a resnet18 with and without pretraining.
05-metadata.ipynb - checking the usability of the metadata in training the model.
06_multilabel_classification.ipynb - We take a subset of the dataframe that includes the images with bleeds present. We create a new column in the dataframe that includes all the sub categories of bleeds present in an image separated by semicolon. We then train a model on this data and use a threshold of 0.5 to make predictions. The threshold is set to a high value because we want to be conservative with medical data and not predict a bleed unless we are very sure.
07a_diff_archs_resnet101.ipynb - We now try different architectures to improve our model's performance. One way to do so is to use more layers. Hence we try resnet101 after resnet50. It does give a higher accuracy however not so significant.
07b_diff_archs_densenet121.ipynb - Anther good architecture for image classification is densenet. Google it to find more information about this architecture.
07c_diff_archs_alexnet.ipynb - Google to learn more about the alexnet architecture.
08_weight_decay_and_tta.ipynb - In this notebook we introduce weight decay to help our model generalise better.
09_mixed_precision.ipynb - In this notebook we try to reduce the training time by training in half precision for some parts and full precision for the others. This is known as mixed precision training
10_progressive_resizing.ipynb - In this notebook we train a model with image size of 64, after a few epochs we train it on a dataset of 128 and finally after few more epochs we train it with images of the full size (256,256). This technique has proved to give better results for CV applications
11_model_deployment.ipynb - We deploy the model in this using an API. The API is seeme.ai.
[2] Qure.ai blog
[5] fastai—A Layered API for Deep Learning
[7] Super-Convergence: Very Fast Training of Neural Networks Using Large Learning Rates
[8] Bag of Tricks for Image Classification with Convolutional Neural Networks
2. Basic EDA + Data Visualization
3. See like a radiologist with systematic windowing