Once the students have sequenced their samples (or using the datasets provided with this report) they could try to identify the origin of their plasmid samples by using BLAST, identifying antibiotic resistance genes (http:https://ardb.cbcb.umd.edu/blast/) and read alignment against the P. Fluorescens gacS gene (e.g. using the alignment workflow described in the assembly tutorial).
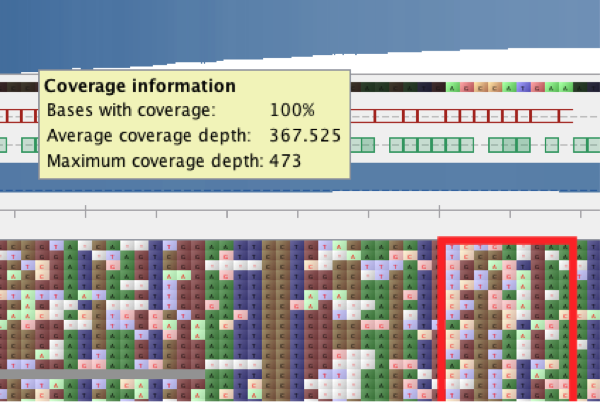
For example, to identify the gacS H294R variant I searched for the relevant wild-type sequence 5’ -AGCCATGAA-3’ in my alignments against the gacS gene (Figure 2).
Figure 1 Pile-up at position 294.
The pile-up shows high variability in this region and largely agrees with the mutated sequences 5’-TCTCGAGAA-‘3 as described by Zuber et al. 1. Thus, it is relatively safe to assume that this sample corresponds to the gacS H294R mutant.
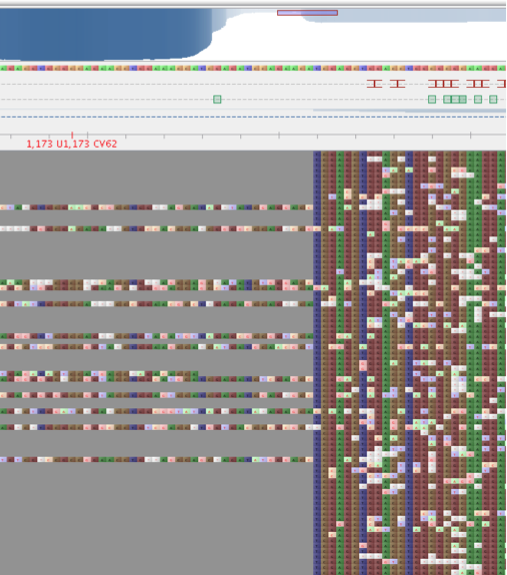
Similarly, the students can search for the Δ76 mutation. In this mutations a 384-bp segment in gacS (128 amino acids located between Met-33 and Leu- 162) were deleted. Note, that during the cloning of this mutant an artificial HindIII site was introduced to allow for excision of this segment. Unaware of this at the time, I used HindIII to linearise the plasmids. It worked well in all of the plasmids apart from one. As we now know, this was the gacS Δ76 mutant (Figure 2).
Figure 2 Alignment of reads against the Δ76 gacS gene. The HindIII incision is clearly visible.
Furthermore, the students can confirm the identity of the plasmids by aligning them to their plasmid backbones pME6010 (P1-2) and pUC18 (P3-P5), respectively.
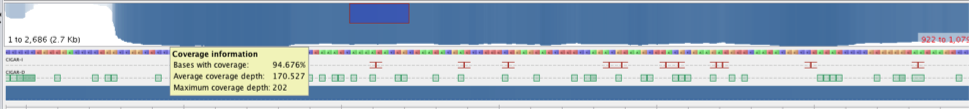 Figure 3 Alignment of reads against pUC18 backbone. The red arrow highlights a ~200 base gap in the pile-up.
Figure 3 Alignment of reads against pUC18 backbone. The red arrow highlights a ~200 base gap in the pile-up.
Note, that while 100% of the bases in pME6010 were covered by reads, only ~95% of the bases in the pUC18 plasmids were covered (see example in Figure 3). Further investigation should be conducted to identify a better backbone reference sequence.