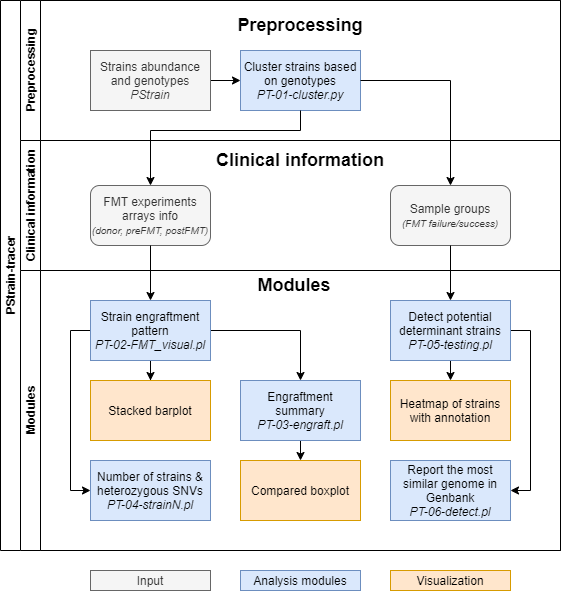
PStrain-tracer is a downstream analysis and visulization package of PStrain, a tool to infer the genome sequence and propotion of strains from whole genome shotgun metagenomics data.
PStrain-tracer was written in python3 and perl5, and using R code to do the visualiztion.
git clone https://github.com/deepomicslab/PStrain-tracer.git
- Python Dependencies
- numpy
- Configuration
- modify the
config.listin thesrc/folder
- modify the
Rscript=/where/R/installed/bin/Rscript
R_lib=/where/R/library
MUMmer=/where/MUMmer/install/MUMmer3.23/
fasttree=/where/excutable/FastTree
ass_sum=/where/PStrain-tracer/src/assembly_summary_refseq_20200423.txt
species_marker=/where/PStrain/db/species_markers.txt.gz
marker_gene=/where/PStrain/db/marker_gene.fna.gz
If you don't use the module of report the most similar strains from Genbank, you only need modify Rscript and R_lib.
- The R_lib should contains:
- ggplot2
- RColorBrewer
- pheatmap
- For the report the similar strains modules, you need:
- MUMmer
- FastTree2
- Assembly Summary from NCBI Assemebly FTP.
- or you can use the
assembly_summary_refseq_20200423.txtwe provided insrcfolder, but remember gunzip before use. species markerandmarker geneare provided in PStrain.
We provide an example to run the package in example folder. example.sh showed the commands used in a compeleted work.
After your installation and configuration. You can change your direction to the example folder and directly run example.sh to figure out if it worked well.
# decompressed the demo PStrain inputs
cat demo_PStrain_result.tar.gz.* | tar -zxv
# Cluster strains based on genotypes
python ../src/PT-01-cluster.py -c config -o demo_PStrain_result --similarity 0.9
# Create the output folder
mkdir output
# Strain engraftment pattern and visualization
perl ../src/PT-02-FMT_visual.pl -WDR output -LS experimets_list.tsv -PS demo_PStrain_result/ -S 0.9
# Engrafment summary and visualization, default is run with proportion, with '-m abd' will run with original input abundance
perl ../src/PT-03-engraft.pl -WDR output -LS experimets_list.tsv -PS demo_PStrain_result/ -S 0.9
perl ../src/PT-03-engraft.pl -WDR output -LS experimets_list.tsv -PS demo_PStrain_result/ -S 0.9 -m abd
# Number of strains and heterozygous SNVs statistic
perl ../src/PT-04-heterzygousSNV_strainN.pl -WDR output -PS demo_PStrain_result/ -S 0.9
# Detect potential determinant strains with wilcox testing
perl ../src/PT-05-wilcox_testing.pl -WDR output -PS demo_PStrain_result/ -S 0.9 -LS group_info.tsv
# Reporting the most similar genome of the provided strains
perl ../src/PT-06-detect.pl -WDR output -PS demo_PStrain_result/ -S 0.9 -LS strain.txt
- config files, the format you can find at PStrain
- experiments list, a line contrains all samples involved in one FMT experiment follow a sequence of [donor] [recipient] [preFMT1] [preFMT2] ... (separated by tab). An example you could find at
example/experimets_list.tsv
FMT30 FMT103 FMT109
FMT34 FMT101 FMT102 FMT110
FMT33 FMT96 FMT97 FMT98 FMT106
FMT2 FMT51 FMT53
FMT1 FMT5 FMT6 FMT39 FMT41
FMT27 FMT75 FMT77
FMT4 FMT66 FMT67
- group info, contrains all samples you want to compare in two groups, one line per sample with sample name and group name, the hander line is needed. An example you could find at
example/group_info.tsv
sampleID groupname
FMT1 Success
FMT2 Failure
FMT27 Failure
FMT30 Success
FMT33 Success
FMT34 Success
FMT4 Failure
- strains list, the strains you interested in with the format you run after the step1. An example you could find at
example/strain.txt
Ruminococcus_lactaris_clu-1
Eubacterium_rectale_clu-4
- PT-01
A new folder namemerge_[similarity_cutoff]will created in the PStrain output folder. Containsstrain_number.txtandseq/. The folderseq/had sequence and abundance of each strains after clustering. - PT-02
A new folder nameoutdir/FMT_visual/will created. For each experimet, it had four files:*.xlsStrain abundace of all samples in this FMT experiment*.rela.vis.txtVisualization input with strain engraftment pattern annotation of each strain*.rela.RVisualization script in R format*.rela.pdfVisualization result in stacked barplot
- PT-03
A new folderoutdir/engraft/will created. For each experimet, it had three files:*.[propotion/relative].tsvVisualization input*.[propotion/relative].RVisualization script in R format*.[propotion/relative].pdfVisualization result in compared boxplot
- PT-04
A new folderoutdir/strainNwill created.strainN_heterSNV_stat.*.tsvThe number of strains number and the corresponding heterozygous SNVs in each species of every samples
- PT-05
A new folderoutdir/wilcox_testing/will created. For each comparison, the result had nine files:*.tsvModified abundance input for strain and species level*.group_infoA copy of group info input for further check*.RWilcox-tesing R script*.test.tsvWilcox-testing result for strain and species level*heatmap*Visualization files
- PT-06
A new folderoutdir/find_strainwill created. Foreach strain, a new folderoutdir/find_strain/[strain_name]will created.
After you run the PT-06 module, you can find three shell inoutdir/find_strain/[strain_name], you need to run them in order.
The separated shell scripts are made in order to avoid the error in downloading files or calling SNP. Finally, you could find result:dl/Contains downloaded genome fastas in the same species of your interested strain.snp/Contains snps calling result of genomestree.nwkThe phylogenetic tree of interested strains and all strains in the sample species in newick format for further visualization or check.
- Packages
Python Bio
Fasttree
MUMmer
wget https://data.gtdb.ecogenomic.org/releases/release207/207.0/bac120_metadata_r207.tar.gz
tar -zxvf bac120_metadata_r207.tar.gz
mv src/
Download and uncompressed, move to src/
You should use v207 as the same version MetaPhlAn used in SGB2GTDB.
- Generate shell
#For MetaPhlAn3
perl ../src/PT-07-detect.v2.pl -WDR test_M3 -S s__Escherichia_coli -V M3 -I s__Escherichia_coli_seq.txt -N 50 -DBS mpa_v31_CHOCOPhlAn_201901.species_markers.txt.gz -DBM mpa_v31_CHOCOPhlAn_201901.fna.bz2
#For MetaPhlAn4
perl ../src/PT-07-detect.v2.pl -WDR test_M4 -S s__Escherichia_coli -V M4 -I s__Escherichia_coli_seq.txt -N 20 -DBS mpa_vOct22_CHOCOPhlAnSGB_202403.species_markers.txt.gz -DBM mpa_vOct22_CHOCOPhlAnSGB_202403.fna
Then will generate shell scripts in test_M*/find_strain/s__Escherichia_coli.
- Run shell scripts step by step for downloading genomes, detecting SNPs and constructing tree to find the closest genomes for each strain.
-IPStrain output of strain sequence e.g. result/seq/s__Escherichia_coli_seq.txt.
Example:
# Gene Locus Ref Alt str-1 str-2 str-3
562__P64549__K758_00320 30 G T 0 1 1
562__P64549__K758_00320 72 A G 1 1 1
562__P64549__K758_00320 140 G C 1 1 1
562__P64549__K758_00320 196 T C 1 1 1
562__P64549__K758_00320 204 A G 1 1 1
562__P64549__K758_00320 210 T C 1 1 1
562__P64549__K758_00320 213 C G 1 1 1
562__P64549__K758_00320 222 A G 1 1 1
562__P64549__K758_00320 246 T C 1 1 1
-DBSSpecies markers file of MetaPhlAn database. Generated using translate_pkl.py from .pkl file in MetaPhlAn database.
Example: python translate_pkl.py mpa_vOct22_CHOCOPhlAnSGB_202403.pkl mpa_vOct22_CHOCOPhlAnSGB_202403.species_markers.txt
-DBMMarker genes fasta file from MetaPhlAn database. Download and uncompressed from MetaPhlAn. Files in .bz2 or .fna or .gz are acceptable.- [option]
-NThe threshold N of genome number. When the genomes in the corresponding species > N, we will randomly select N genomes to construct tree.
- tree.nwk A tree of selected genomes and input strain in newick format.
- *sorted_distance.txt For each strain, have an output with genomes in incremental distance.
Example:
s__Escherichia_coli_str-1 0.0
GCF_003303795.1_ASM330379v1 0.268918215
GCF_002459435.1_ASM245943v1 0.273527672
GCF_015316785.1_ASM1531678v1 0.273704976
GCF_005395845.1_ASM539584v1 0.27497622299999996
GCF_003334035.1_ASM333403v1 0.27671494
Jiang, Yiqi, et al. "A framework to trace microbial engraftment at the strain level during fecal microbiota transplantation." bioRxiv (2022).