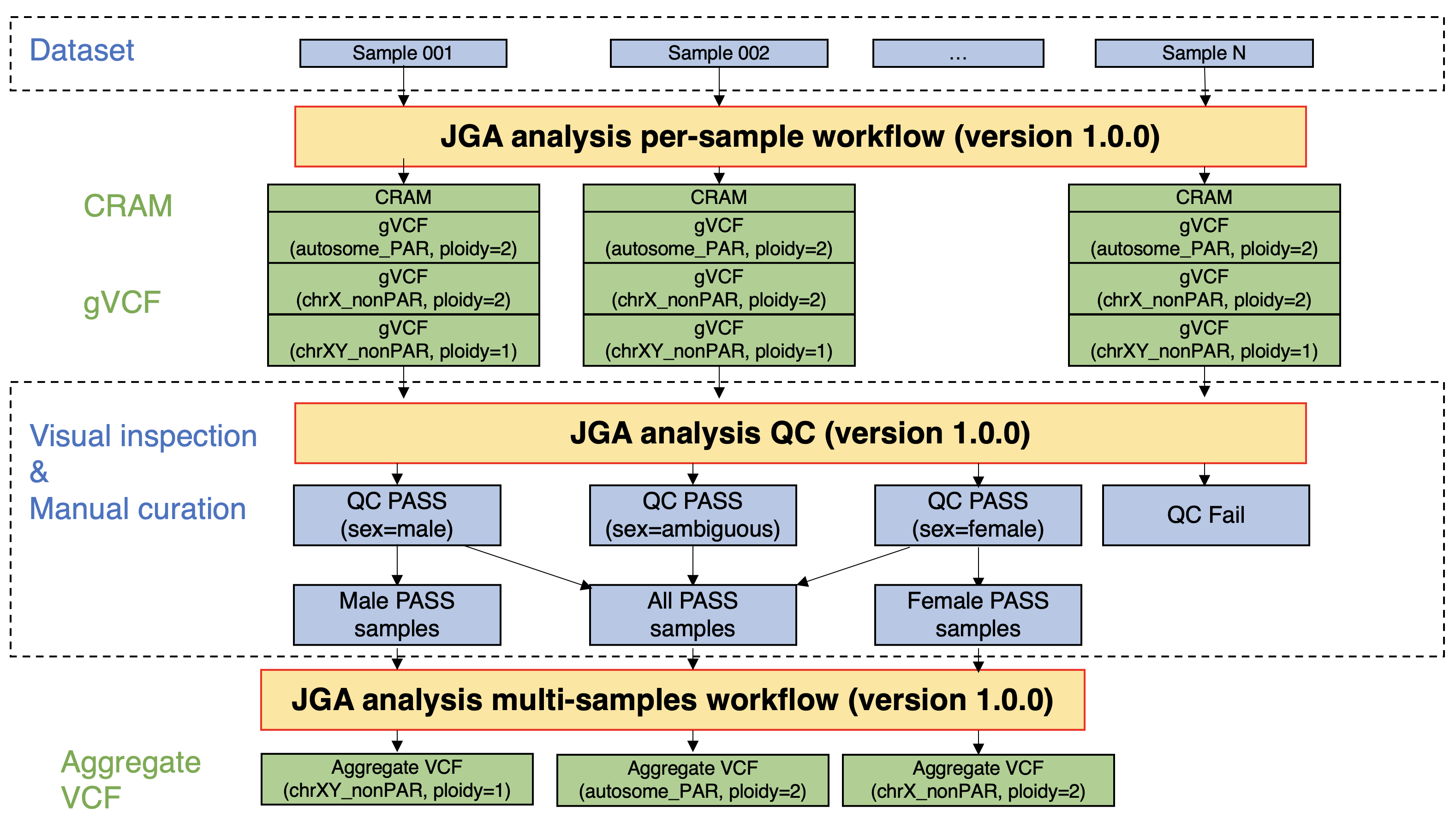
In Japanese Genotype-phenotype Archive (JGA), most of the whole-genome sequencing (WGS) data are registered in the FASTQ format. Accordingly, the data users have to download the WGS data, followed by data processing by themselves. To improve the convenience of the data users, germline WGS data registered in JGA were processed in a certain workflow, and alignment results (CRAM), variant call results per sample (gVCF), and variant call results per dataset (aggregated VCF) were calculated. The post-processing data have been registered in the JGA, and the data users can download the post-processing data from the JGA.
- JGA analysis per-sample workflow. This workflow takes FASTQ files as input, aligns them to the reference genome (GRCh38), and performs variant call per sample. The alignment results (CRAM), variant call results per sample (gVCF), and quality control metrics (CRAM-level and gVCF-level metrics) used in later steps are output.
- JGA analysis QC. This program performs quality control (QC) by visualizing the cram- and gVCF-level metrics calculated by the abovementioned JGA analysis per-sample workflow.
- JGA analysis multi-samples workflow. This workflow takes multiple gVCF files as input, performs joint call and variant quality score recalibration (VQSR), and outputs variant call results per dataset (aggregate VCF). The summarized data (sites-only aggregate VCF) was then calculated.
The JGA analysis per-sample workflow can be executed with the JGA analysis job manager.