This script prints input_hr.dat hamiltonian from the wannier90 package to block-divided form between orbitals.
python HOPPING.py input out.dat
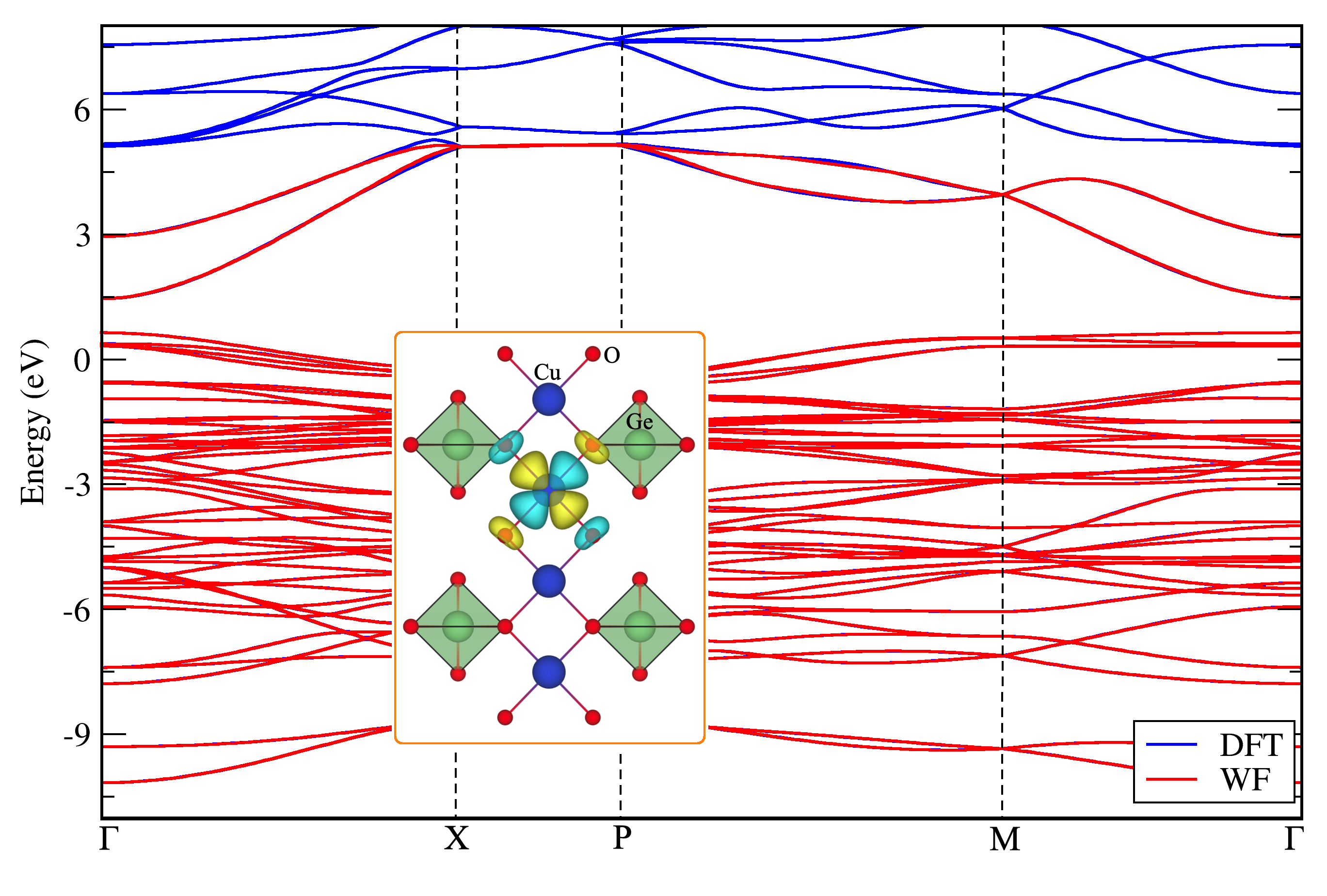
As an example let's consider PBE band structure of Cu2GeO4 system [Phys. Rev. B 100, 214401 (2019)], which was parametrized by Wannier functions based on Cu(d), O(p) and Ge(s) initial projectors. Total number of Wannier functions is 46, corresponding to 20(5x4) d orbitals of copper, 24(3x8) p orbitals of oxygen and 2(1x2) s orbitals of germanium atoms:
input.json file contains the following information:
"cell_vectors": [[5.596748, 0.000000, 0.000000], [0.000000, 5.596748, 0.000000], [2.798374, 2.798374, 4.700648]],
"number_of_atoms": 14,
"max_sphere_num": 10,
"print_complex": 0,
"positions": [[2.79837, 4.19756, 1.17516], [6.99593, 5.59675, 3.52549], [2.79837, 1.39919, 1.17516], [4.19756, 5.59675, 3.52549], [5.59675, 6.94556, 3.37506], [1.44956, 5.59675, 1.02474], [5.59675, 4.24793, 3.37506], [4.14719, 5.59675, 1.02474], [2.79837, 4.24793, 3.67591], [1.44956, 2.79837, 1.32558], [2.79837, 6.94556, 3.67591], [4.14719, 2.79837, 1.32558], [0.00000, 0.00000, 0.00000], [5.59675, 2.79837, 2.35032]],
"wanniers": [5,5,5,5,3,3,3,3,3,3,3,3,1,1]- cell_vectors - (3x3)(dfloat) array of unit cell vectors (in Ang);
- number_of_atoms - (int) total number of atoms, parametrized by Wannier functions;
- positions - (3 x number_of_atoms)(dfloat) array of cartesian coordinates of these atoms (in Ang);
- wanniers - (number_of_magnetic_atoms) (int) array of Wannier functions numbers of these atoms;
- max_sphere_num - (int) maximum number of coordination sphere to print the hamiltonian blocks between pair of atoms;
- print_complex - (0 or 1) print the imaginary part of hamiltonian blocks.
As a result, one can find out.dat file, which contains the information from input.json file and output data:
==================================================================
Hopping <a|H|b> between atom 0 (000)<-->atom 0 ( 0 0 0 ) in sphere # 0 with radius 0.000000 -- 1:
Radius vector is: 0.000000 0.000000 0.000000
Hopping <a|H|b> between atom 0 (000)<-->atom 5 ( 0 0 0 ) in sphere # 1 with radius 1.949268 -- 1:
Radius vector is: -1.348810 1.399190 -0.150420
...
==================================================================
For example, the last table gives the hamiltonian block 5x3 <Cu(d)|H|O(p)> between atom 0 (Cu(d)) and atom 5 (O(p)) with the distance between them 1.949268 Å and radius vector R = (-1.348810, 1.399190, -0.150420)Å.