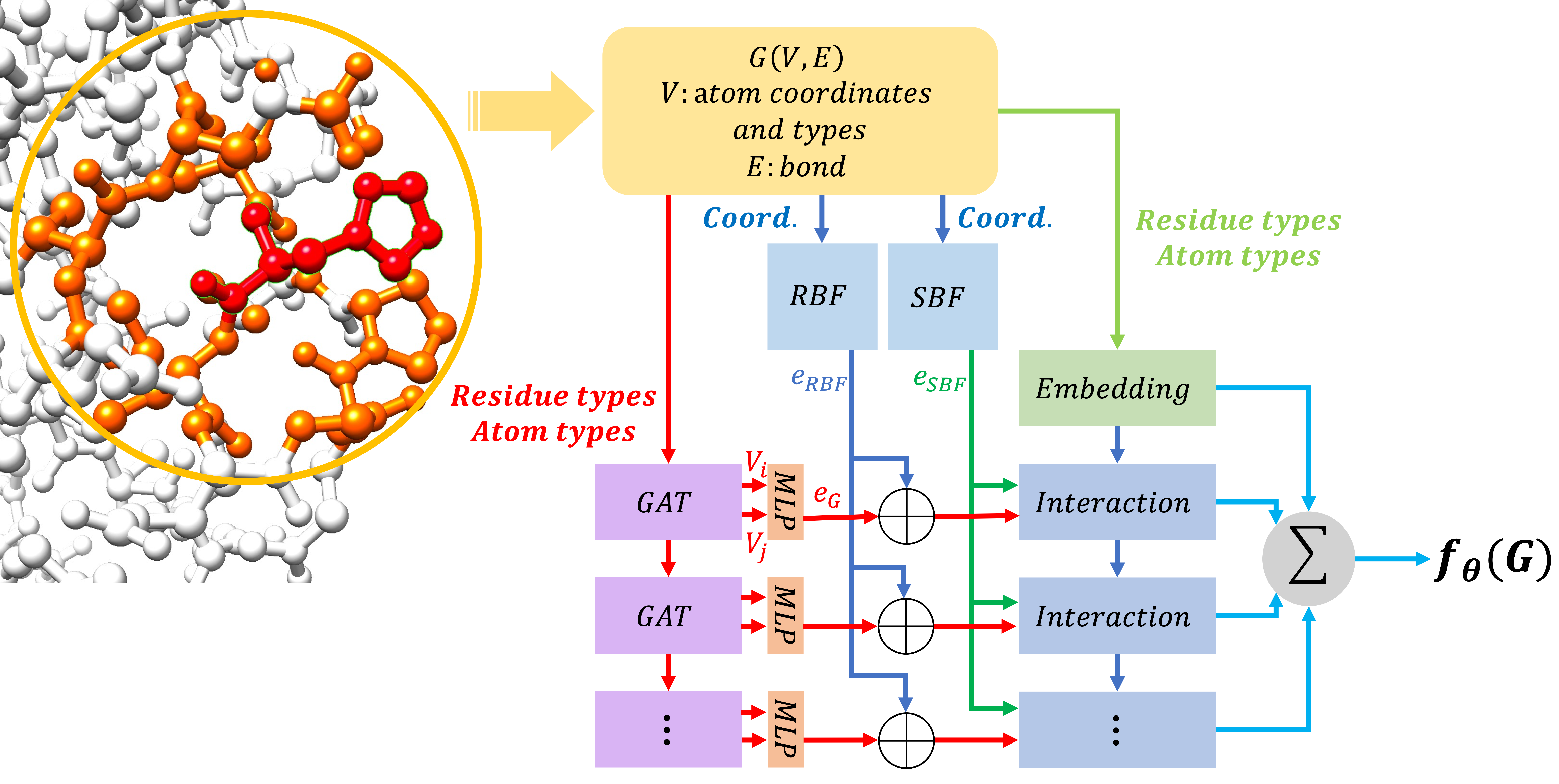
A graph attention network as an energy-based model for protein conformation.
The following step is required in order to implement GraphEBM:
To install the dependencies for the project, execute
conda env create -f environment.yaml
Following the https://github.com/facebookresearch/protein-ebm#downloading-the-datasets and https://github.com/facebookresearch/protein-ebm#preprocessing to get the datasets.
The following script will train a model:
python train.py \
--model DimeNetPlusPlusGraph \
--max-distance 20.0 \
--neg-sample 3 \
--multisample 16 \
--n-epochs 2 \
--gmm \
--uniform \
--no-augment \
--gpus 1 \
--nodes 1 \
--batch-size 16 \
--nodes 1 \
--data-workers 0 \
--exp DimeNetPlusPlusGraph3Sample20DistSmooth0.75 \
--log-interval 50 \
--save-interval 2000 \
--resume-iter 0 \
--smooth_factor 0.75 \
--no-restrict
In our paper, we trained models on a RTX3090.
To reproduce the results shown in our paper, run
# Discrete sampling strategy
python vis_sandbox.py \
--exp=DimeNetPlusPlusGraph3Sample20DistSmooth0.75 \
--resume-iter=24000 \
--task=rotamer_trial \
--sample-mode=gmm \
--neg-sample=150
# Countiuous sampling strategy
python vis_sandbox.py \
--exp=DimeNetPlusPlusGraph3Sample20DistSmooth0.75 \
--resume-iter=24000 \
--task=rotamer_trial \
--sample-mode=rosetta \
--neg-sample=500
In our paper, the trained model has been uploaded in https://drive.google.com/file/d/1nhJL0Cj53BmK5dRJqXSkKn-klBuvh2v2/view?usp=sharing. You can download it and put it in cachedir/DimeNetPlusPlusGraph3Sample20DistSmooth0.75.
If you find the GraphEBM useful, please cite:
@article {Liu2022.09.05.506704,
author = {Liu, Deqin and Chen, Sheng and Zheng, Shuangjia and Zhang, Sen and Yang, Yuedong},
title = {SE(3) Equivalent Graph Attention Network as an Energy-Based Model for Protein Side Chain Conformation},
elocation-id = {2022.09.05.506704},
year = {2022},
doi = {10.1101/2022.09.05.506704},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2022/09/06/2022.09.05.506704},
eprint = {https://www.biorxiv.org/content/early/2022/09/06/2022.09.05.506704.full.pdf},
journal = {bioRxiv}
}