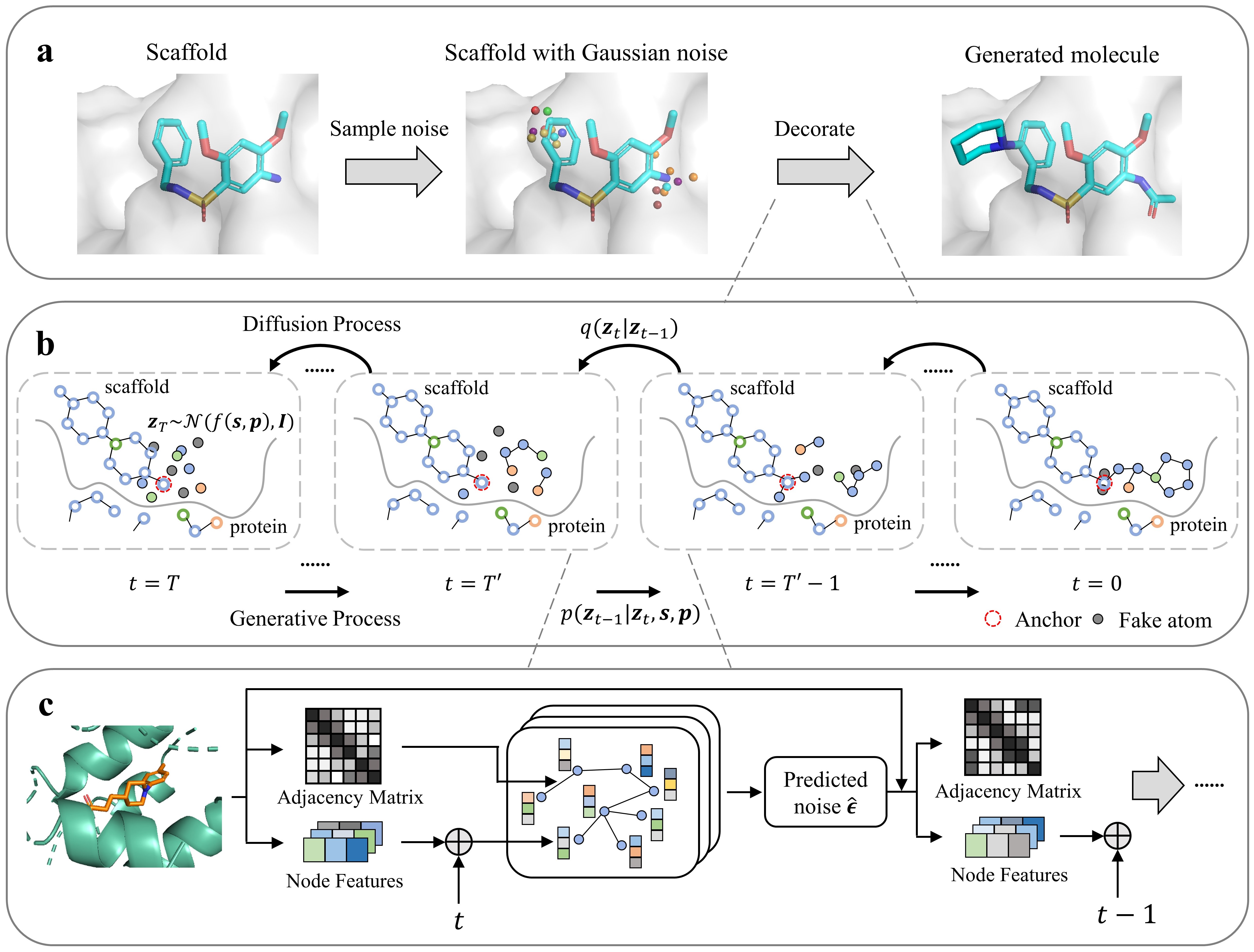
DiffDec is an end-to-end E(3)-equivariant diffusion model to optimize molecules through molecular scaffold decoration conditioned on the 3D protein pocket.
conda env create -f environment.yamlPlease refer to README.md in the data folder.
To train a model for single R-group decoration task, run:
python train_single.py --config configs/single.ymlTo train a model for multi R-groups decoration task, run:
python train_multi.py --config configs/multi.ymlYou can sample 100 decorated compounds for each input scaffold and protein pocket and change the corresponding parameters in the script. You can also download the model checkpoint file from this link and save it into ckpt/. Run the following:
bash sample.shYou will get .xyz and .sdf files of the decorated compounds in the directory sample_mols.
You can run evaluation scripts after sampling decorated molecules:
bash evaluate.shTo generate R-groups for your own pocket and scaffold, you need to provide the pdb structure file of the protein pocket, the sdf file of the scaffold, and the scaffold's smiles with anchor(s). For Example:
CUDA_VISIBLE_DEVICES=0 python sample_single_for_specific_context.py --scaffold_smiles_file ./data/examples/scaf.smi --protein_file ./data/examples/protein.pdb --scaffold_file ./data/examples/scaf.sdf --task_name exp --data_dir ./data/examples --checkpoint ./ckpt/diffdec_single.ckpt --samples_dir samples_exp --n_samples 1 --device cuda:0