This repository contains code to reproduce all figures from our paper:
Fulcher, B. D., Murray, J. D., Zerbi, V., & Wang, X.-J. (2019). Multimodal gradients across mouse cortex. PNAS, 201814144.
You will need to fill the Data directory with some processed data that you can download from this figshare repository.
This paper would not have been possible without the generous sharing of open data (thank you!). Please consult each original data source for detailed licensing information for each dataset.
Allen Institute gene-expression data was retrieved using custom code that queries the Allen SDK.
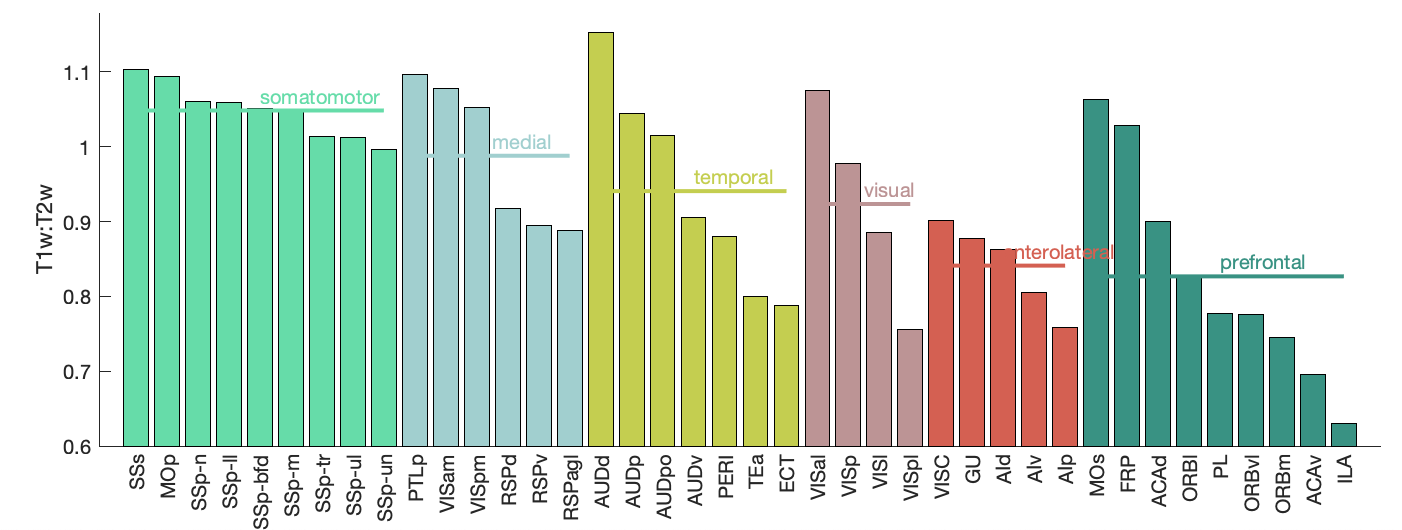
Figure 1A generated manually mouse-brain imaging software (by Valerio Zerbi).
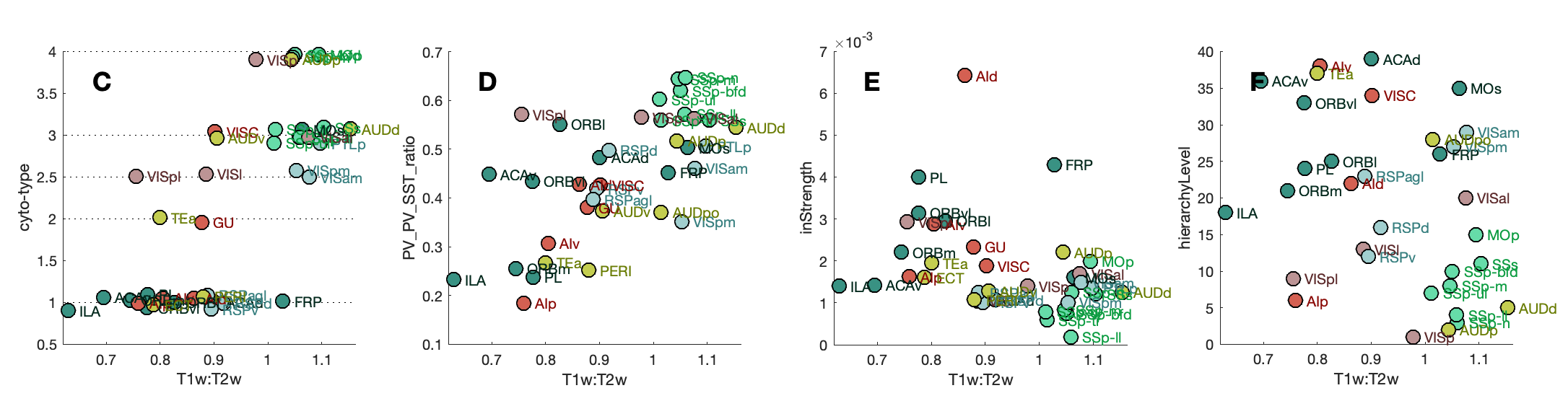
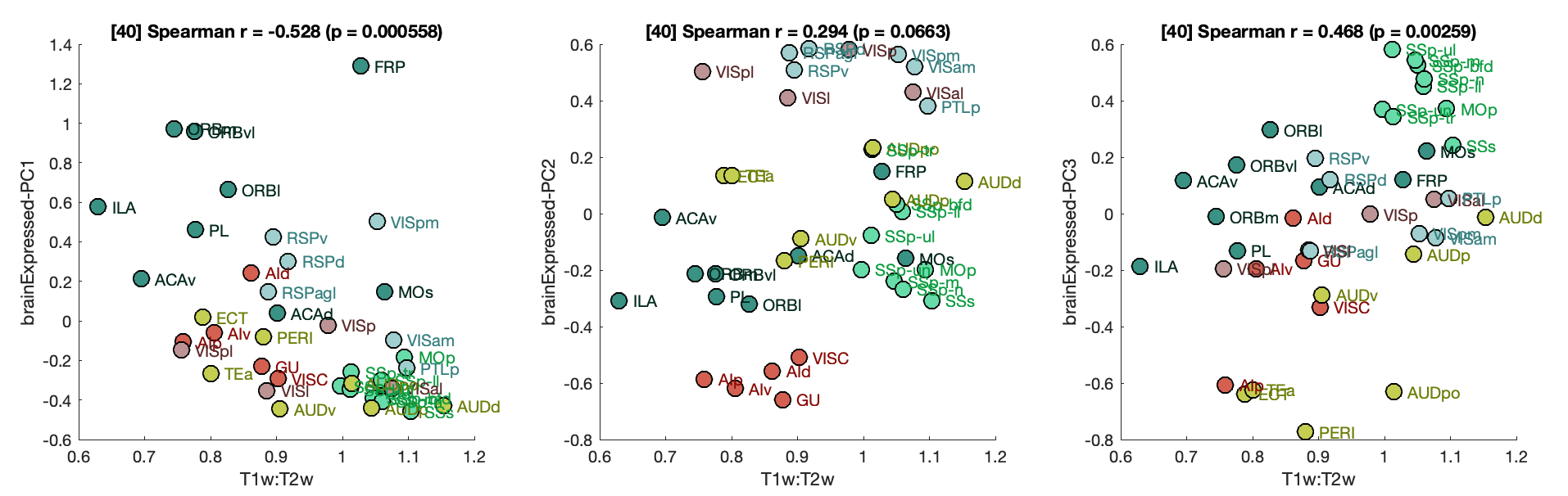
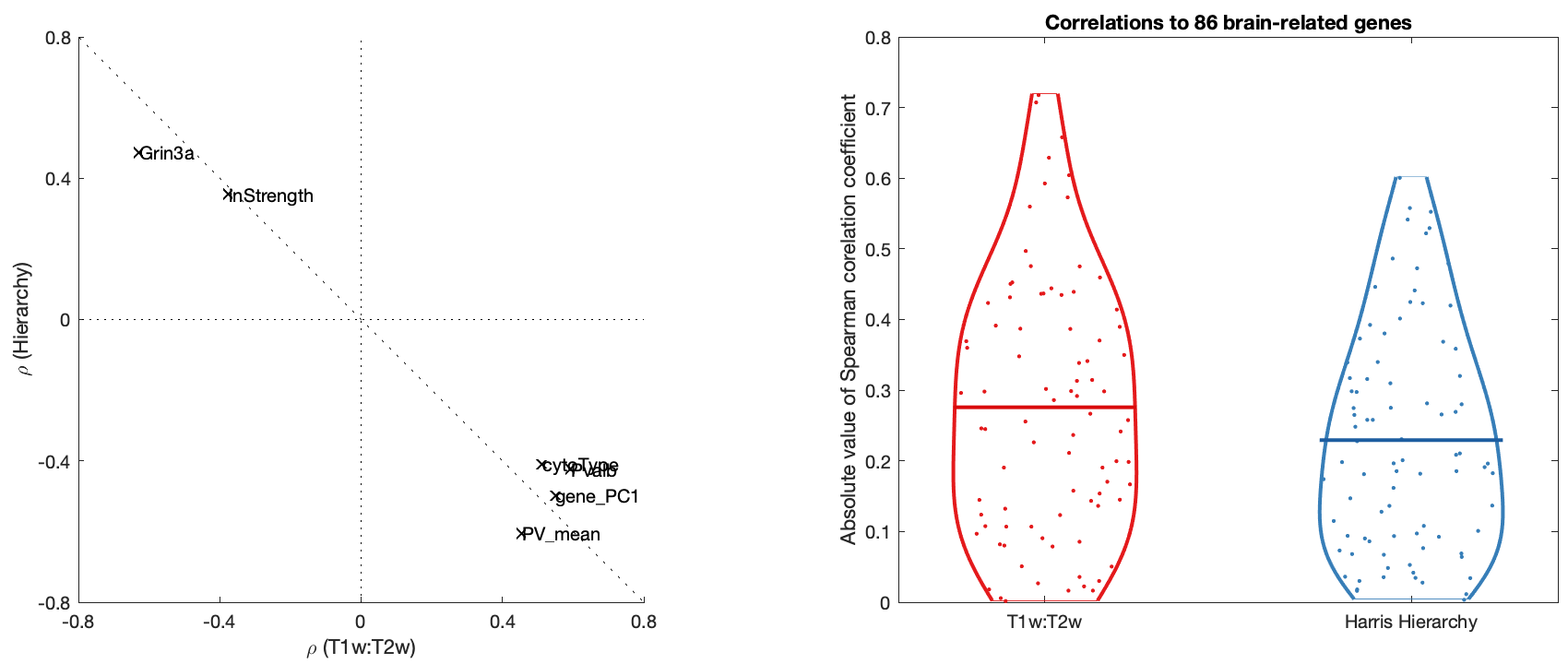
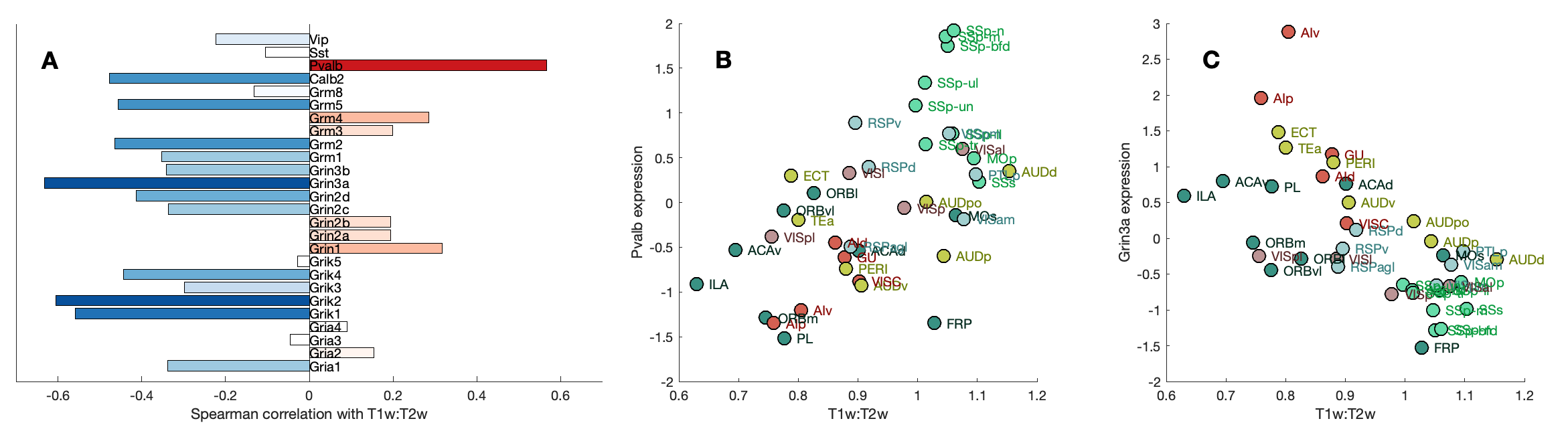
FunctionalClasses()T1T2Corrs()SpecificGeneFigure()Note that B was removed from the final paper; which only plots the Grin3a scatter
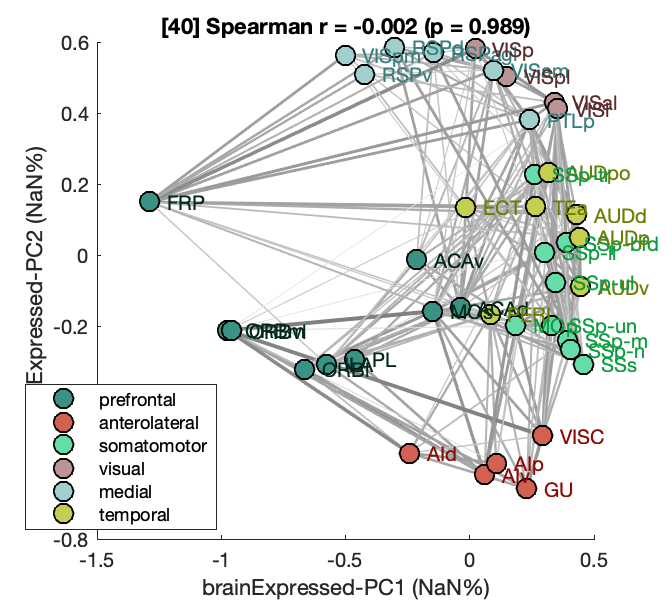
MassAction('brainExpressed','scaledSigmoid','benCombo','bayesian')Note that additional outputs are also produced, visualizing analytics about the principal components, including Fig.S4.
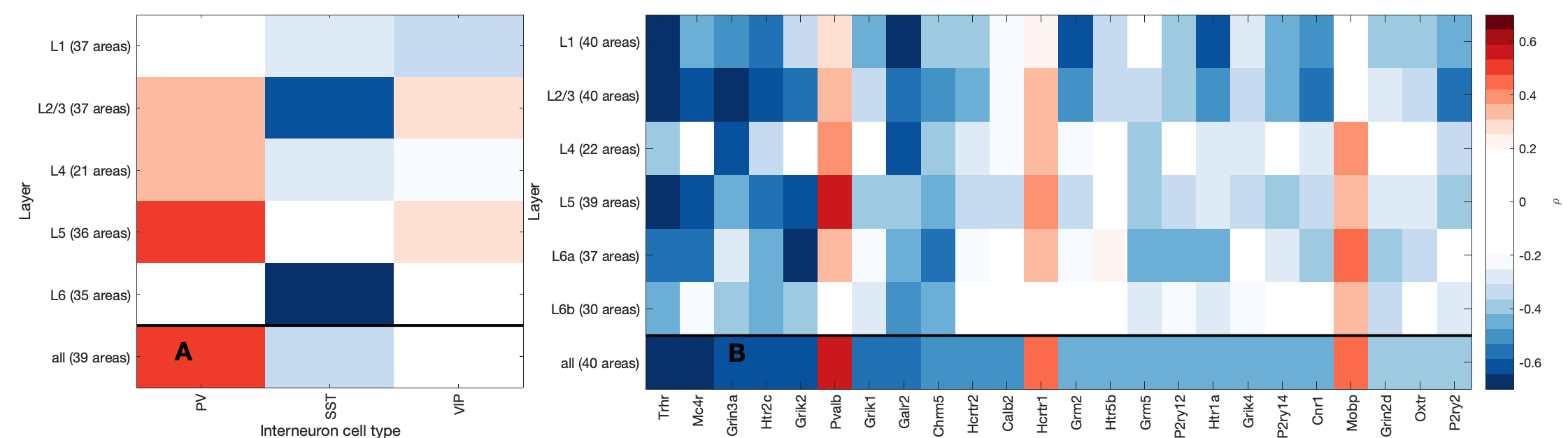
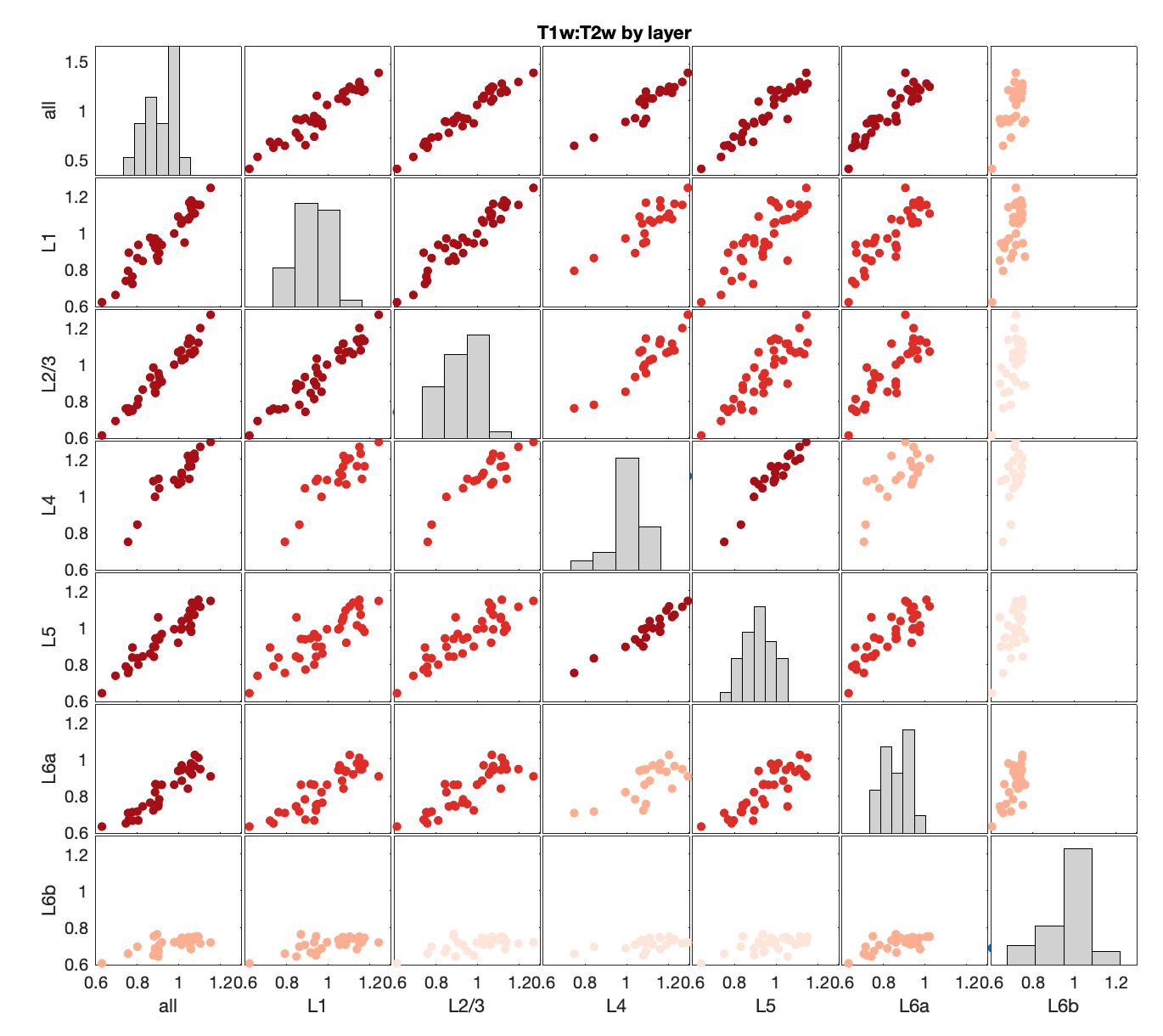
LaminarPlot()DiverseMatrixT1T2()
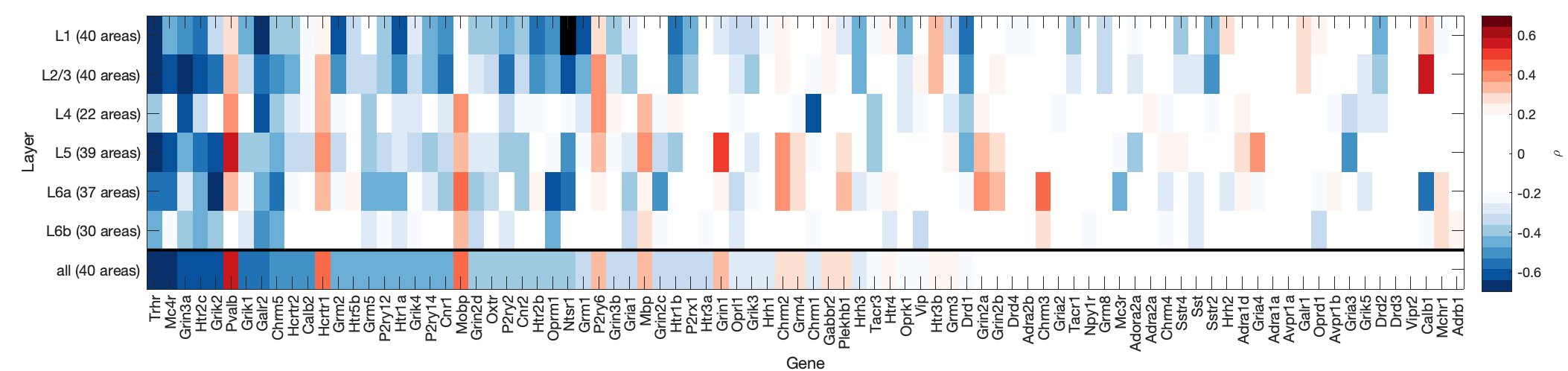
PlotBigDamnMatrix()G = LoadMeG('cortexAll');
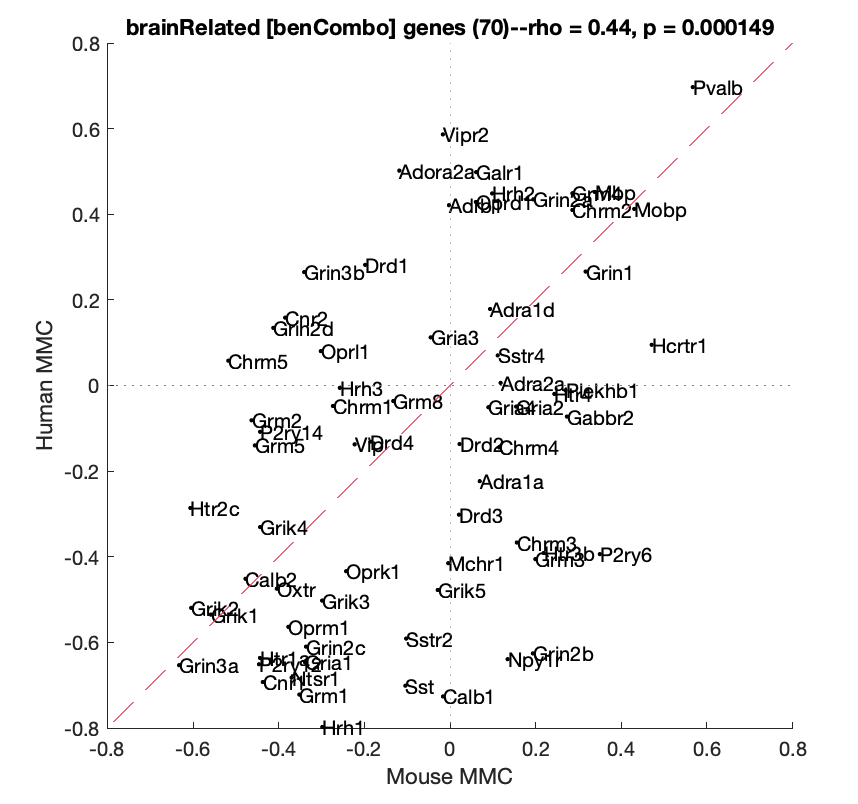
HumanMouseComparisonAnal(G,'brainRelated','benCombo',true)Figs S1, S2, S3 generated manually using mouse-brain imaging software (by Valerio Zerbi).
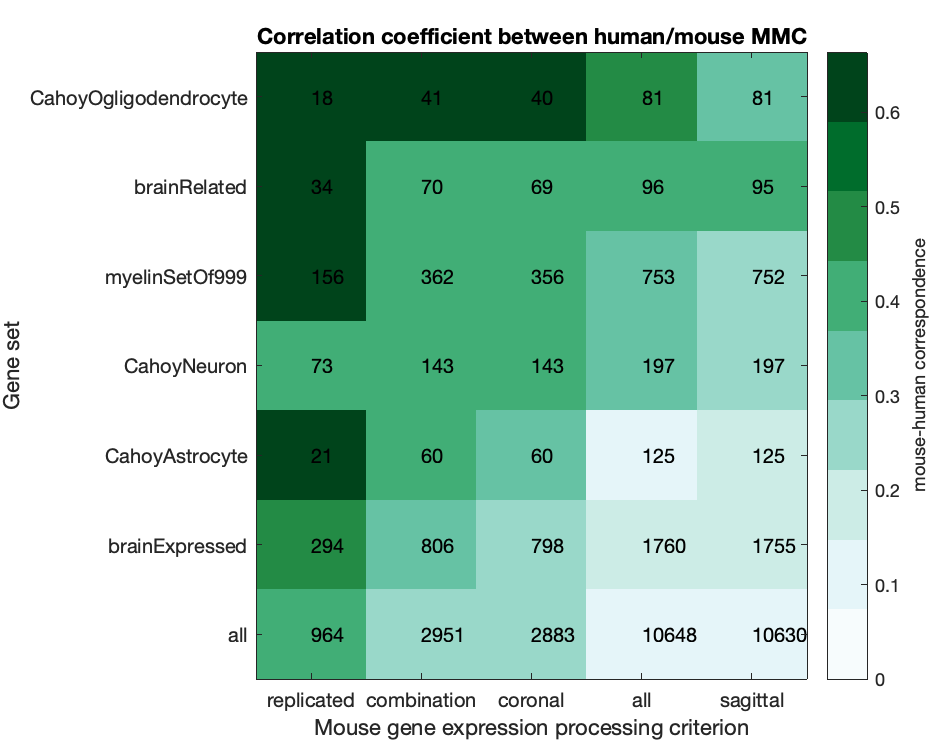
MassAction('brainExpressed','scaledSigmoid','benCombo','bayesian')T1T2ByLayerMyelinCorrs('CellAtlas')TheBestLayers('genes',true,[]);T1T2_or_hierarchyFigure S9: Comparison of mouse-human correspondence as a function of expression processing and gene-set filtering
whatGeneSets = {'all','brainExpressed','brainRelated','CahoyNeuron','CahoyOgligodendrocyte','CahoyAstrocyte','myelinSetOf999'};
whatSectionFilters = {'sagittal','coronal','combZ','benCombo','replicated'};
HumanMouseSectionCompare(whatGeneSets,whatSectionFilters);