For a python notebook with an example of how one might use pp-netlib standalone, see tests/example/mlst_example.
$ git clone https://github.com/bacpop/pp-netlib.git
$ cd pp-netlib
$ python setup.py install
You can check that things are working as they should by running bash tests/run_tests.sh.
from pp_netlib.network import Network
example_graph = Network(ref_list = samples_list, outdir = "path/to/output/dir", backend = "backend")
## construct from a list of edges
example_graph.construct(network_data = edges_data_df, weights = weights)
## load from file (.gt, .graphml, .xml)
example_loaded_network = Network([], outdir = "path/to/output/dir", backend = "backend")
example_loaded_network.load_network("path/to/network_file.graphml")
## produce an mst with cluster-plot and stress-plot images
example_graph.visualize(viz_type = "mst", out_prefix = "some_prefix")
## produce a bunch of output files to load into cytoscape, including merging metadata
example_graph.visualize(viz_type = "cytoscape", out_prefix = "some_prefix", external_data="path/to/metadata.csv")
## prune graph with 4 threads, make sure "type_isolate_id" is retained in the pruned graph
example_graph.prune(type_isolate = "type_isolate_id", threads = 4)
## add new edges from new_data, with specified new vertex labels
example_graph.add_to_network(new_data_df = new_data, new_vertex_labels = ["new_label_1", "new_label_2", "new_label_3"])
## get summary stats
example_graph.get_summary() #summary written to stderr, no text file produced
example_graph.get_summary("summary_file.txt") #summary written to stderr AND to summary_file.txt
## save graph to file, also save pruned graph if it exists in the Network instance
example_graph.save(file_name="example_graph", file_format=".graphml", to_save="both")
## write node labels, clusters, and metadata to some_prefix_node_data.tsv, edge data to some_prefix_edge_data.tsv
example_graph.write_metadata(out_prefix = "some_prefix")
PopPUNK is a tool for population partitioning using nucleotide kmers. More specifically, it first generates variable-length kmers from a set of input genome assemblies. Then, it creates kmer-sketches -- subsets of the ordered kmer set for each assembly -- which it uses to calculate pairwise distances between all the input assemblies (this is the database-creation step). It then applies a user-specified statistical model to these all-vs-all pairwise distances to yeild clusters, each of which represents a strain in the population.
Networks are useful representations of these pairwise distances. In the context of PopPUNK, each node in a network represents an assembly. In the initial all-vs-all pairwise state, every node in the PopPUNK network is connected to every other node via edges that are weighted with the corresponding pairwise distance. Once a model is applied, some of the edges in this graph are discarded based on their associated weights, and what remains is a multi-component graph where each component, or cluster, is a distinct strain, as mentioned above.
The purpose of pp-netlib is to take these graph functions and wrap them in a separate python module, so that it can be imported into PopPUNK where needed, and also so that it can be used standalone for similar applications.
Whenever a graph is created in a Network instance, whether you load or construct one, it is passed through a function called prepare_graph. This function ensures that all nodes in the graph have a node label, and a component membership attribute (i.e. a cluster identity attribute). It also check whether edges are weighted. If a list of labels is passed to the function, then it uses those, otherwise, it names nodes as node1, node2, node3 and so on by default. Similarly, the function can be passed a python dictionary with nodes as the keys and the component they belong to, as corresponding values. If such a dictionary is not passed (for example when constructing a graph), component membership is calculated and stored as a node attribute.
Graph-tool is quite fast as its core functionalities are written in C++, but when adding nodes to a graph, it uses (and expects the inputs to be in the form of) integers. That is, nodes are identified by an integer, and new nodes added to existing networks of size n, get identities of n+1, n+2 and so on. In order to preserve uniformity across graph-tool and NetworkX (wherein nodes can be arbitrary Python data types), pp-netlib also maps node labels in the input data to integers. First, it cleans up the labels, removing some special characters as noted elsewhere, and sorts the resulting list. Next, it creates an integer mapping to each node label. Finally, it converts the node labels in the edge data to their corresponding integer maps.
Thus, edge data that looks like this:
source target
a b
b c
b d
c e
Becomes:
source target
0 1
1 2
1 3
2 4
And a dictionary {"a":0, "b":1, "c":2, "d":3, "e":4} is stored as an attribute of the Network instance. When .add_to_network is called, pp-netlib checks whether any of the new vertex labels already exist in the graph, and removes them from the new_vertex_labels list. Then it updates the vertex map, preserving the mappings for pre-existing nodes, and appending mappings for new ones. Thus, adding ["a", "f", "g"] to a Network instance would result in the following updated dictionary: {"a":0, "b":1, "c":2, "d":3, "e":4, "f":5, "g":6}.
One distinct advantage of PopPUNK is that adding new samples to one of these premade models is very quick. The tool achieves this by using clique-pruning on the model network to create a reference graph. Essentially, the tool iterates over all cliques in the model network and retains one node from each, ultimately reducing the number of nodes in the reference graph and therefore the number of pairwise comparisons that need to be made in order to assign a new sample to a precalculated strain.
Another thing to note about pruning is that when using NetworkX, a known issue is that pruning is not deterministic. Since reference nodes from each clique are chosen at random, the exact identity of the set of reference nodes, and therefore the number and identity of the resulting edges changes when NetworkX is used. This is not an issue when using graph-tool however.
example_graph.outdirexample_graph.backendexample_graph.ref_listexample_graph.graph(initialised as None; can be used directly as a graph-tool or networkx graph if called in this way)example_graph.ref_graph(initialised as None, only exists if pruning is done)example_graph.mst_network(initialised as None, only exists if visualisation withviz_type="mst"is done)
example_graph.vertex_labels(cleaned and sortedref_list)example_graph.vertex_map(see note)example_graph.weights
example_graph.edge_dataexample_graph.sample_metadata
example_graph = Network(ref_list, outdir, backend)
ref_list (list), required: List (or set) of sample labels (str), one for each node. Any special characters (".", ":", ")", "(" ) in the labels are replaced with underscores.
NOTE: When loading a graph, a ref_list value is still expected; an empty list will work.
outdir (pathlike/str), optional: Path (ideally absolute path) to where any outputs will be written. Defaults to "./", i.e. the working directory.
backend (str), optional: Graph analysis library to use. Accepted values are "GT" and "NX", for graph-tool and NetworkX. A backend can be set this way (i.e. specifying as an argument to the Network instance) or as an environment variable.
NOTE: If you do both of the above, the value passed as an arg to the instance will take precedence.
The latter can be achieved as follows:
## set as env variable inside of a python script that contains Network instance
os.environ["GRAPH_BACKEND"] = "GT"
*OR*
## set as bash/shell env variable
export GRAPH_BACKEND=GT ## inside bash/shell terminal/environment
## construct from a list of edges
example_graph.construct(network_data, weights)
network_data (pd.DataFrame *OR* list *OR* scipy.sparse.coo_matrix), required: Data containing a series of edges. This can be in one of the three data types noted.
- If a dataframe is passed, it is expected to have a column called "source" and a column called "target". Optionally, it can have a column called "weights".
- If a list is passed, it is expected to be a list of tuples where the first element of each tuple is the source node and the second is the target node. Optionally, each tuple can contain a third element, which will be used as the weight for that edge.
## weighted list
edge_list = [(s1, t1, w1), (s2, t2, w2), (s3, t3, w3)...]
## unweighted list
edge_list = [(s1, t1), (s2, t2), (s3, t3)...]
- If a sparse matrix (arbitrarily called "edge_matrix") is provided, edge_matrix.row should return source nodes, edge_matrix.col should return destination nodes, and edge_matrix.data should return weights
weights (list *OR* bool), optional: A list of weights to apply to the edges. These are expected to be in the same order as the edges in network_data. Defaults to None.
- If
network_datais a dataframe and contains weights (in a column called "weights") already,weightscan be set to True. Note that in this case, if there is no column called "weights", an error is thrown. If weights exists innetwork_databut should not be used, it can be set to False. Ifweightsis not specified, an unweighted network will be constructed regardless of whethernetwork_datacontains a "weights" column. - If
network_datais an unweighted list, andweightsis also a list, a weighted Network is constructed. Ifweightsis set to False in this case, an unweighted Network is made. However, if an unweighted list andweights=Trueis passed, an error will be thrown. If a weighted list is passed,weightsneeds to be set to True. - If
network_datais a sparse matrix, theweightsargument is ignored and a weighted network will be created.
## load from file (.gt, .graphml, .xml)
## note that initialisation is different, an empty list is passed to `ref_list`
example_loaded_network = Network(ref_list = [], outdir, backend)
example_loaded_network.load_network(network_file, sample_metadata_csv)
network_file (pathlike/str), required: Path (ideally the absolute path) to the graph file you'd like to load into the Network instance.
sample_metadata_csv (pathlike/str), optional: Path (ideally the absolute path) to a file containing node metadata. This is useful if the graph you'd like to load in has no node attributes already saved in the network_file. The program looks for a "Taxon" or "sample_id" column, and a "Cluster" column in sample_metadata_csv, and uses these data to apply node labels and cluster assignments.
NOTE: Loading in ".gt" files will only work with backend = "GT". ".xml" and ".graphml" files can be used by either backend.
example_graph.visualize(viz_type, out_prefix, external_data)
viz_type (str), required: The type of visualisation to produce. Accepted values are "mst" and "cytoscape".
- If "mst" is specified, a new "mst" directory, within the Network instance
outdiris created, and the following outputs are written:out_prefix_mst_network_data.graphml (graphml file of the minimum spanning tree)out_prefix_mst_cluster_plot.png (left)out_prefix_mst_stress_plot.png (right)
| Cluster Plot | Stress Plot |
|---|---|
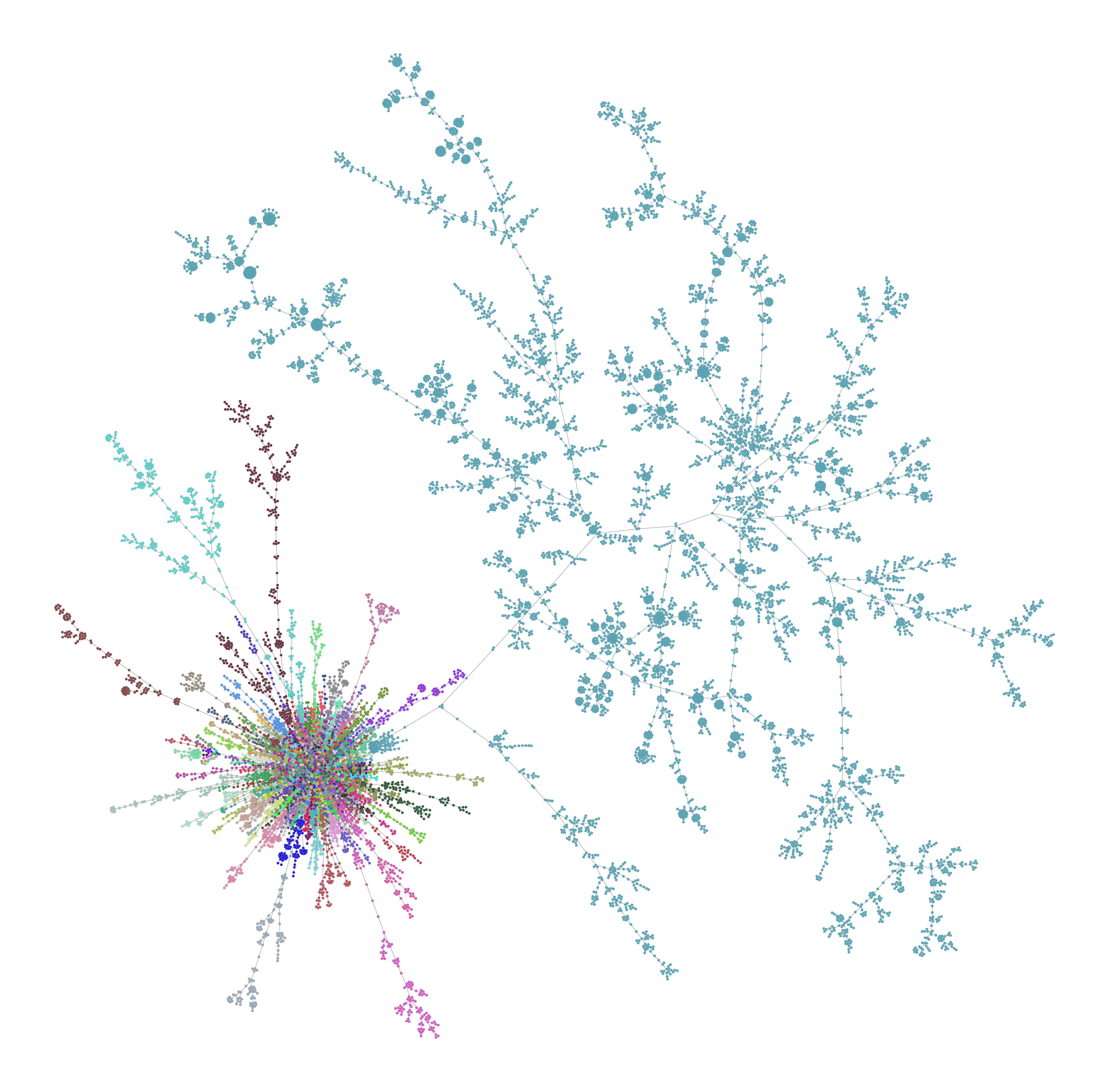 |
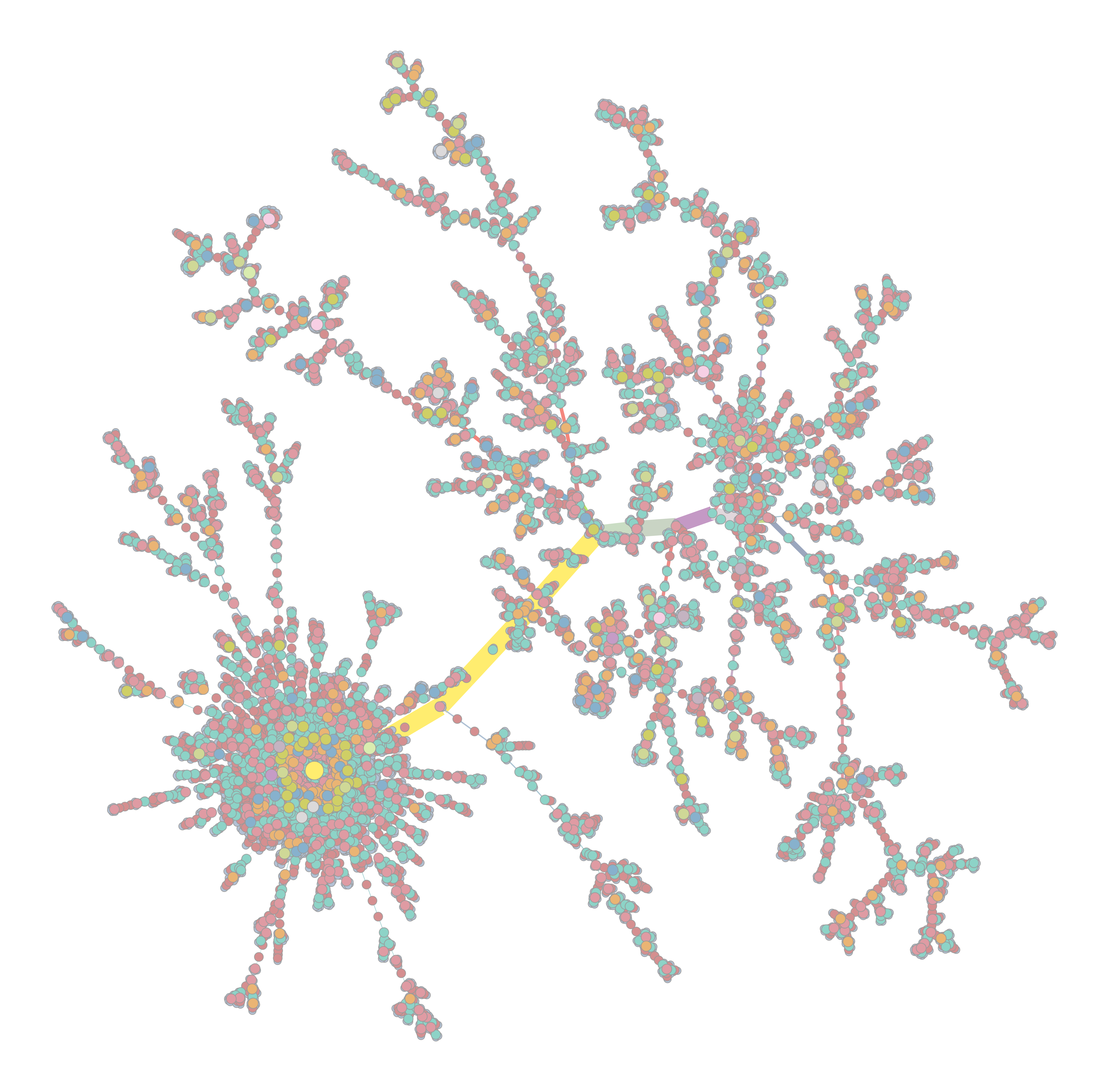 |
- If "cytoscape" is specified, a new "cytoscape" directory is created as above and the following outputs are generated:
out_prefix_mst.graphml (this is produced ONLY if the Network instance contains an MST, i.e. isviz_type="mst"has been run first)out_prefix_cytoscape.graphml (graphml file of the full graph)out_prefix_component_1.graphml throughout_prefix_component_N.graphml (each graph component is individually saved)out_prefix_node_data.tsv (sample metadata)out_prefix_edge_data.tsv
## prune graph
example_graph.prune(type_isolate, threads)
type_isolate (str), optional: The sample label associated with the type isolate in the data. Once pruning is done, if type_isolate was not retained, it is added back into the graph. A type isolate is chosen by PopPUNK. Defaults to None.
threads (int), optional: The number of threads to use: pruning can be time-consuming for very big graphs. Defaults to 4.
## add new edges from new_data, with specified new vertex labels
example_graph.add_to_network(new_data_df, new_vertex_labels)
new_data_df (pd.DataFrame), required: A dataframe containing the new edges to add. It is expected to contain a "source" column and a "target" column.
- If you are adding data to a weighted graph, the program looks for a "weights" column in
new_data_df, and if no such column is found, arbitrarily weights all new edges as 0. - If you are adding to an unweighted graph, then the new edges will also be added unweighted.
new_vertex_labels (list), required: A list of all unique node labels in new_data_df.
## get summary stats
example_graph.get_summary(print_to_std, summary_file_prefix)
print_to_std (bool) optional: Whether to print network summary statistics to stderr. Defaults to True.
summary_file_prefix (str), optional: Whether to write network summary statistics to a text file. Defaults to None.
## save graph to file, also save pruned graph if it exists in the Network instance
example_graph.save(file_name, file_format, to_save)
file_name (str), required: Prefix to be applied to the saved file.
file_format (str), required: The file format to use. Accepted values are ".gt" (only when backend = "GT"), ".graphml", and ".xml".
to_save (str), optional: Whether to save only the full graph (to_save="full_graph"), only the pruned graph (to_save="ref_graph"), or to save both (to_save="both"). If "both" is given but the graph was not pruned, only the full graph is saved.
NOTE: Visualising with viz_type = "cytoscape" saves the full graph to file as noted above.
## write node labels, clusters, and metadata to some_prefix_node_data.tsv, edge data to some_prefix_edge_data.tsv
example_graph.write_metadata(out_prefix, meta_outdir, external_data)
out_prefix (str), required: Prefix to be applied to all metadata files.
meta_outdir (pathlike/str), optional: A separate directory to save metadata files to. Defaults to the outdir of the Network instance.
external_data (pathlike/str *OR* pd.DataFrame), optional: A file containing other data associated with each sample.
Writing metadata produces:
- out_prefix_node_data.tsv (with external_data merged by sample_id)
- out_prefix_edge_data.tsv`