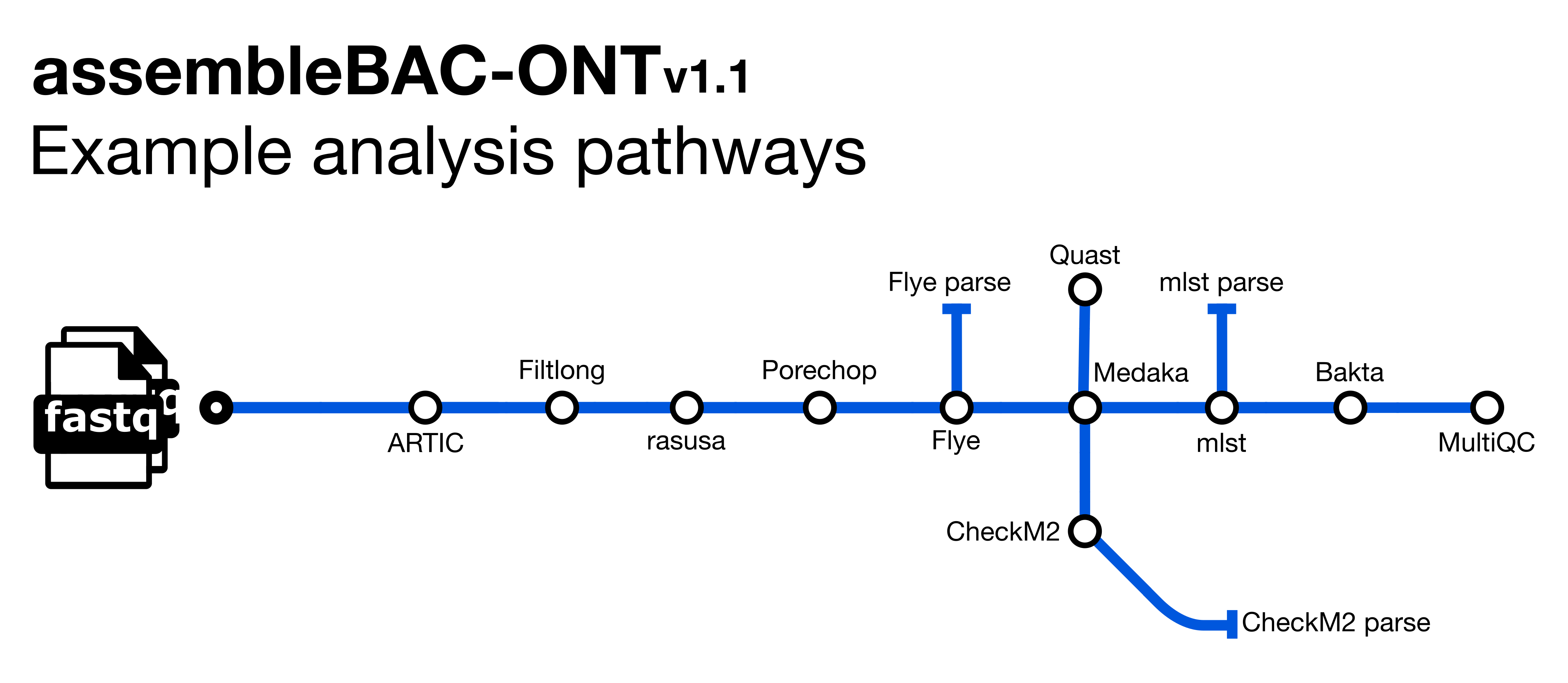
avantonder/assembleBAC-ONT is a bioinformatics pipeline that de novo assembles and annotates Oxford Nanopore (ONT) long-read sequence data.
- Aggregate pre-demultiplexed reads from MinKNOW/Guppy (
artic guppyplex) - Filter long reads (
Filtlong) - Downsample fastq files (
Rasusa) - Remove adapter sequences (
Porechop) - de novo assembly (
Flye) - Polish assemblies with ONT data (
Medaka) - Assembly metrics (
Quast) - Assembly completeness (
CheckM2) - Sequence Type assignment (
mlst) - Annotation (
Bakta) - Present QC, visualisation and custom reporting for filtering and assembly results (
MultiQC)
-
Install
nextflow(>=23.04.0) -
Install any of
Docker,Singularity,Podman,ShifterorCharliecloudfor full pipeline reproducibility (please only useCondaas a last resort; see docs) -
Download the
Baktalight database (Baktais required to run theamrfinder_updatecommand):wget https://zenodo.org/record/7669534/files/db-light.tar.gz tar -xzf db-light.tar.gz rm db-light.tar.gz amrfinder_update --force_update --database db-light/amrfinderplus-db/
-
Download the
CheckM2database (CheckM2is required):checkm2 database --download --path path/to/checkm2db
-
Start running your own analysis!
- Typical command for assembly and annotation
nextflow run avantonder/assembleBAC-ONT \ -profile singularity \ -c <INSTITUTION>.config \ --input samplesheet.csv \ --fastq_dir path/to/fastq/files \ --genome_size <ESTIMATED GENOME SIZE e.g. 4M> \ --medaka_model <MEDAKA MODEL> \ --min_read_length <MINIMUM READ LENGTH> \ --outdir <OUTDIR> \ --baktadb path/to/baktadb/dir \ --checkm2db path/to/checkm2db/dir/uniref100.KO.1.dmnd \ -resume
See usage docs for all of the available options when running the pipeline.
Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. This is usually done in the form of a config profile (<INSTITUTION>.config in the example command above). You can chain multiple config profiles in a comma-separated string.
- The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker.- Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment.- If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs.- If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
The avantonder/assembleBAC-ONT pipeline comes with documentation about the pipeline usage, parameters and output.
avantonder/assembleBAC-ONT was originally written by Andries van Tonder. I wouldn't have been able to write this pipeline with out the tools, documentation, pipelines and modules made available by the fantastic nf-core community. In particular, the excellent viralrecon pipeline was a source of code and inspiration. Finally, a shout out to Robert Petit's Dragonflye as an additional source of inspiration.
If you have any issues, questions or suggestions for improving assembleBAC-ONT, please submit them to the Issue Tracker.
If you use the avantonder/assembleBAC-ONT pipeline, please cite it using the following doi: ZENODO_DOI
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.