Learning to Discover Crystallographic Structures with Generative Adversarial Networks
This repository is a TensorFlow implementation of CrystalGAN : CrystalGAN: Learning to Discover Crystallographic Structures with Generative Adversarial Networks. AAAI Spring Symposium: Combining Machine Learning with Knowledge Engineering 2019
- Python 2.7
- Jupyter
- Tensorflow-gpu
- Scikit-learn
- Tqdm
- Pymatgen
- Matplotlib
- Numpy/Scipy
Note: Experiments must be run using GPU with powerful graphics card.
Clone the repository with:
git clone https://github.com/asmanouira/CrystalGAN
Then:
cd CrystalGAN/
CrystalGAN is based on three steps:
- First and second steps are implemented in
CrystalGAN_step1+2.py - Third and last step is implemented in
CrystalGAN_step3.py
Launch jupyter notebook:
jupyter notebook
- Open
Step1+Step2_CrystalGAN.ipynb
CrystalGAN takes as input two datasets of binary compounds and generates as output ternary compounds.
We choose in this implementation as example Pd-H "Palladium - Hydrogen" and Ni-H "Nickel - Hydrogen",
The aim is to generate novel ternary compounds of Pd-H-Ni "Palladium - Hydrogen - Nickel"
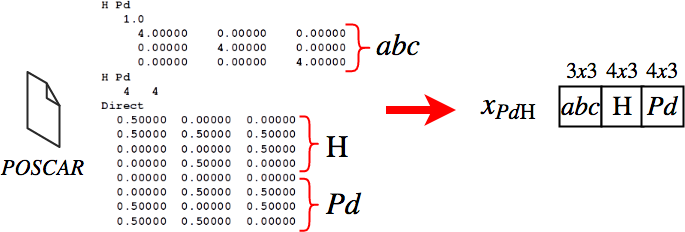
Samples in our datasets are POSCAR files and converted to 4D tensors as shown below:
In order to prepare the inputs for complexity augmentation , we add an empty placeholder for each dataset:
This procedure described above was implemented in Matlab: POSCAR2mat.m
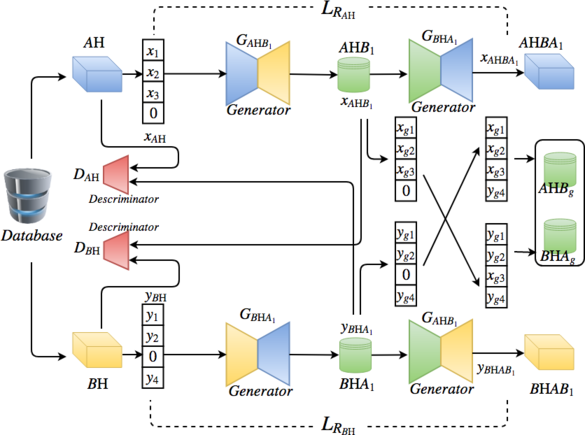
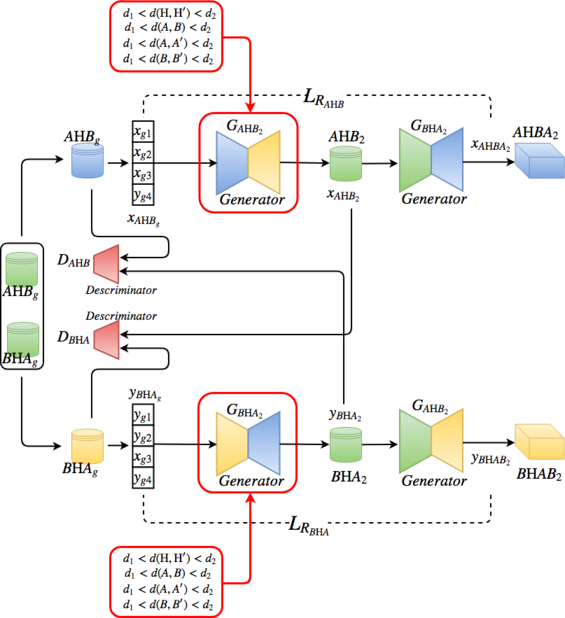
CrystalGAN is composed basically of two cross-domain GANs.
Each encoder and decoder of the generators and the discriminators are composed of fully-connected layers.
The output datasets of the first network (including STEP1 and STEP2) will be trained by the second cross-domain GAN
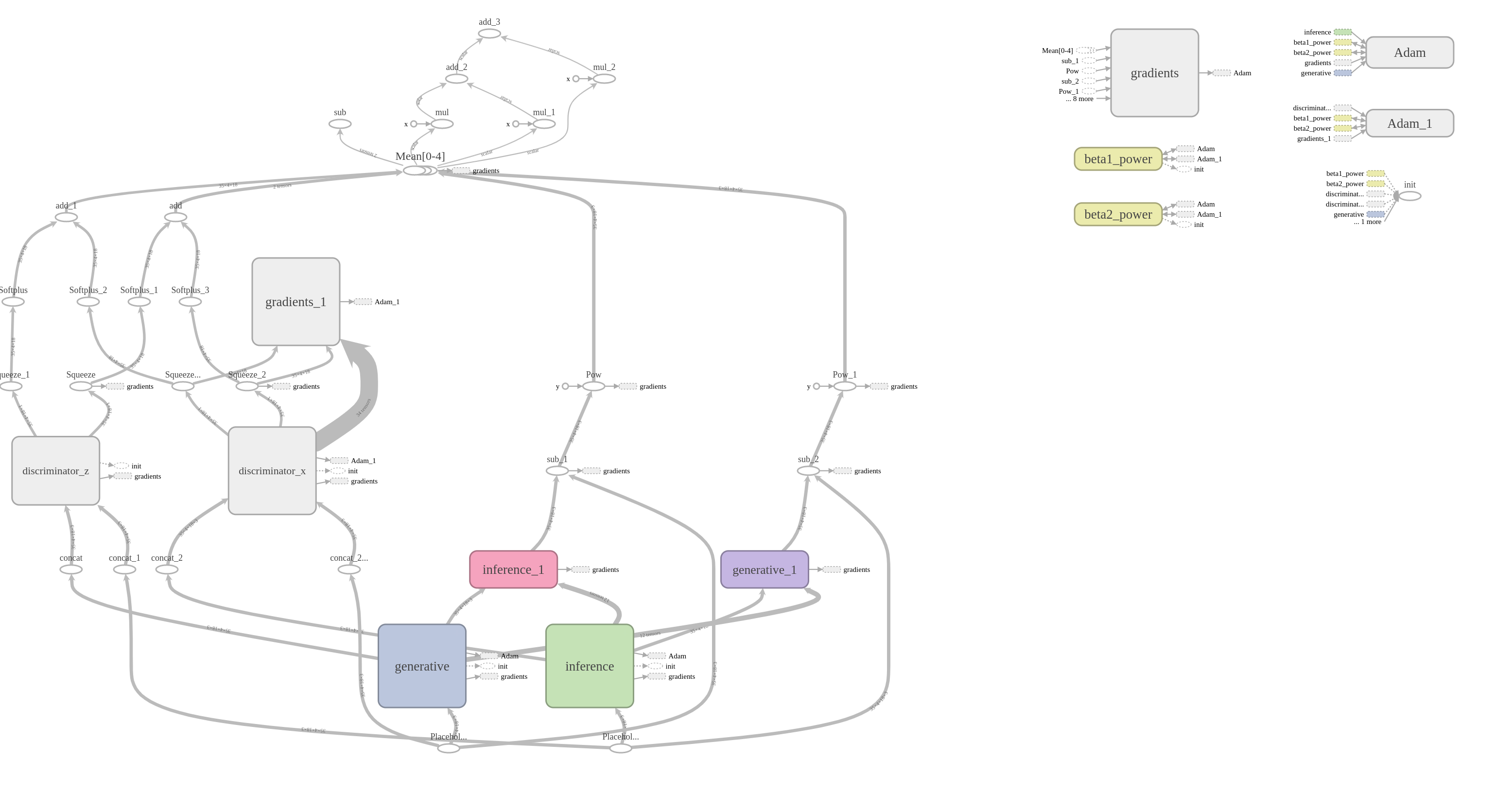
To check the architecture of CrystalGAN network, we can use tensorboard:
tensorboard --logdir=graphs/
CrystalGAN generates ternary compounds in 4D tensors and then print them as POSCAR files. We evaluate the generated crystal structures by:
- Visualizing the lattice of the crystal VESTA using the generated POSCAR files.
- Visualizing their distances histogram of first neighbors for all atoms in the cell.
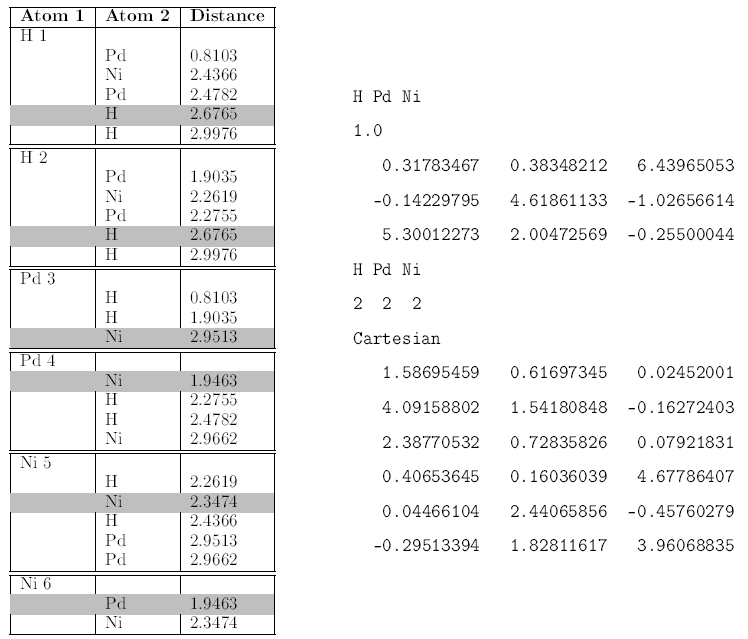
- Check if the first neighbors distances respect the reinforced constraints by printing them in tables
To compute neighbors of all atoms in a crystallographic structure using POSCAR file as input argument: see neighbors.py
An example of a POSCAR file is in data/.
In our study, the penalized first neighbors distances are between the atoms Pd-Pd', Ni-Ni', Pd-Ni and H-H'.
Those distances fixed to be between d1 = 1.8 Å and d2 = 3 Å.