- Install Homebrew
- Install cromwell
brew install cromwell
- Install Docker
https://hub.docker.com/editions/community/docker-ce-desktop-mac
Tools tested:
| Tool | 5mC (CpG) | 5mC (GpC) | 6mA | Notes |
|---|---|---|---|---|
| Nanopolish | X | X | ||
| Dorado | X | ? | (10.4.1 only) | |
| Megalodon | X | X | X | |
| NanoHiMe | X | X | About 8 hours per GB of fast5s |
- NanoHiMe paper HepG2 Hia5/K27me3 fast5.
Local Kraken copy of run 1: /aryeelab/users/mark/NanoHiMeref/HepG2H3K27me3_1/HepG2_nanoHiMe_H3K27me3_1.pod5
- HepG2 CpG methylation (ENCODE WGBS) bigbed
Dorado unmapped bam: - gs:https://fc-bd302969-686c-4e8d-a857-5c5bc13f265e/submissions/ff458a62-88a1-4570-8c5f-589e79c9dc53/dorado_basecall/fe20757f-14f9-487c-9bd7-e8bf6973d8ec/call-basecall/HepG2_nanoHiMe_H3K27me3_1.unmapped.bam - /aryeelab/users/martin/projects/nanopore-benchmarking/dorado/HepG2_nanoHiMe_H3K27me3_1.unmapped.bam
- HepG2 K27me3 (ENCODE ChIP-Seq) bigwig
The commands below are run in a GCP VM to speed up transfer to/from GCP buckets:
Create the VM:
gcloud compute instances create pod5-instance --project=aryeelab --zone=us-central1-a --machine-type=e2-highcpu-16 --network-interface=network-tier=PREMIUM,subnet=default --maintenance-policy=MIGRATE --provisioning-model=STANDARD --service-account=303574531351-compute@developer.gserviceaccount.com --scopes=https://www.googleapis.com/auth/devstorage.read_only,https://www.googleapis.com/auth/logging.write,https://www.googleapis.com/auth/monitoring.write,https://www.googleapis.com/auth/servicecontrol,https://www.googleapis.com/auth/service.management.readonly,https://www.googleapis.com/auth/trace.append --create-disk=auto-delete=yes,boot=yes,device-name=instance-1,image=projects/ubuntu-os-cloud/global/images/ubuntu-2204-jammy-v20230302,mode=rw,size=500,type=projects/aryeelab/zones/us-central1-a/diskTypes/pd-balanced --no-shielded-secure-boot --shielded-vtpm --shielded-integrity-monitoring --labels=ec-src=vm_add-gcloud --reservation-affinity=any
SSH into the VM:
gcloud compute ssh --zone "us-central1-a" "pod5-instance" --project "aryeelab"
Install pod5 tools and screen
sudo apt update
sudo apt -y install python3-pip screen
screen -dR
sudo pip install pod5
Download fast5s from bucket:
# Authenticate with GCP
gcloud auth login
gsutil -m cp -r gs:https://griffin-lab/ONT_data/HEPG2-H3K27me3test .
# Convert a single FAST5 into a (small) POD5
pod5 convert fast5 -t 12 HEPG2-H3K27me3test/FAQ78510_5809c6e8_0.fast5 --output HEPG2-H3K27me3test-small.pod5
# Convert all FAST5s into a (large monolithic) POD5
# Trying to convert all fast5s into a monolithic pod5 directly fails (See https://github.com/nanoporetech/pod5-file-format/issues/33)
time pod5 convert fast5 HEPG2-H3K27me3test/*.fast5 --output pod5s/ --one-to-one ./HEPG2-H3K27me3test/
# Don't do this:
#time pod5 convert fast5 -t 12 HEPG2-H3K27me3test/*.fast5 --output HEPG2-H3K27me3test.pod5
Upload pod5s to bucket:
[TODO: gsutil cp ....]
# Install minimap2 as in https://github.com/aryeelab/nanopore-tools/blob/dev/Docker/minimap2/Dockerfile
# cp minimap2 /usr/local/bin
# Install modbam2bed
sudo apt-get -y install autoconf libbz2-dev liblzma-dev libcurl4-openssl-dev libssl-dev
git clone --recursive https://github.com/epi2me-labs/modbam2bed.git
cd modbam2bed
make modbam2bed
sudo cp modbam2bed /usr/local/bin
# Install bedGraphToBigWig
wget https://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64.v369/bedGraphToBigWig
sudo mv bedGraphToBigWig /usr/local/bin/
# Get genome fasta
gsutil cp gs:https://aryeelab/genome-fasta/Homo_sapiens.GRCh38.dna.primary_assembly.fa .
samtools faidx ${REF}
cut -f1,2 ${REF}.fai > ${REF}.chrom_sizes
SAMPLE="small_test_sample"
# Convert unaligned sam to fastq preserving base modification tags, then pipe into minimap2 for alignment and output an aligned sam file. The y tag carries modification info
REF="Homo_sapiens.GRCh38.dna.primary_assembly.fa"
samtools fastq -T "*" ${SAMPLE}.unmapped.bam | minimap2 -ax map-ont -y $REF - > ${SAMPLE}.sam
# Drop secondary and supplmentary alignments. Sort. Index.
samtools view -bF 0*900 ${SAMPLE}.sam | samtools sort - > ${SAMPLE}.sorted.primary.bam
samtools index ${SAMPLE}.sorted.primary.bam
# Make Bedmethyl file with modification info:
modbam2bed -t 12 -m 5mC --cpg ${REF} ${SAMPLE}.sorted.primary.bam > ${SAMPLE}.5mc_cpg.bed
# Convert BEDmethyl -> Bedgraph -> Bigwig
awk '$10 > 0 {printf "%s\t%d\t%d\t%2.3f\n" , $1,$2,$3,$11}' ${SAMPLE}.5mc_cpg.bed > ${SAMPLE}.5mc_cpg.bedgraph
bedGraphToBigWig ${SAMPLE}.5mc_cpg.bedgraph ${REF}.chrom_sizes ${SAMPLE}.5mc_cpg.bw
- Methylation (5mC & 6mA) call for MinION R10.3 reads #20 For MinION R9.4.1 reads, Megalodon and res_dna_r941_min_modbases-all-context_v001.cfg in rerio worked perfectly!! I'm looking forward to doing same analysis with MinION R10.3 reads.
# "Manually" on a COS GCP machine:
# See https://cloud.google.com/container-optimized-os/docs/how-to/run-gpus
# Start the VM with an NVIDIA A100 40GB GPU from the cloud console (https://console.cloud.google.com/compute/)
# or command line as below. In this case we're naming it `dorado-1`.
gcloud compute instances create dorado-1 --project=aryeelab --zone=us-central1-a --machine-type=a2-highgpu-1g --network-interface=network-tier=PREMIUM,subnet=default --maintenance-policy=TERMINATE --provisioning-model=STANDARD --service-account=303574531351-compute@developer.gserviceaccount.com --scopes=https://www.googleapis.com/auth/devstorage.read_only,https://www.googleapis.com/auth/logging.write,https://www.googleapis.com/auth/monitoring.write,https://www.googleapis.com/auth/servicecontrol,https://www.googleapis.com/auth/service.management.readonly,https://www.googleapis.com/auth/trace.append --accelerator=count=1,type=nvidia-tesla-a100 --create-disk=auto-delete=yes,boot=yes,device-name=dorado-1,image=projects/cos-cloud/global/images/cos-101-17162-127-8,mode=rw,size=200,type=projects/aryeelab/zones/us-central1-a/diskTypes/pd-balanced --no-shielded-secure-boot --shielded-vtpm --shielded-integrity-monitoring --reservation-affinity=any
# SSH into the VM
gcloud compute ssh --zone "us-central1-a" "dorado-1" --project "aryeelab"
# Install NVIDIA driver
sudo cos-extensions list # Current default: 470.161.03
sudo cos-extensions install gpu
# Make the driver installation path executable by re-mounting it.
sudo mount --bind /var/lib/nvidia /var/lib/nvidia
sudo mount -o remount,exec /var/lib/nvidia
# Get FAST5 data
mkdir -p dat/fast5s
toolbox gsutil cp gs:https://aryeelab-joung/nanopore/test/small-sample1/* /media/root/home/$USER/dat/fast5s/
# Start a docker container with access to the FAST5 dir and the GPU
docker run --rm -it \
--volume /home/$USER/dat:/dat \
--volume /var/lib/nvidia/lib64:/usr/local/nvidia/lib64 \
--volume /var/lib/nvidia/bin:/usr/local/nvidia/bin \
--device /dev/nvidia0:/dev/nvidia0 \
--device /dev/nvidia-uvm:/dev/nvidia-uvm \
--device /dev/nvidiactl:/dev/nvidiactl \
nvidia/cuda:12.0.1-devel-ubuntu22.04
# Install basic packages
apt-get update && \
apt-get -y install libbz2-dev \
liblzma-dev \
python3-pip \
samtools \
wget
# Install pod5 tools and duplex tools
pip install pod5 duplex_tools
# Install Dorado
wget -q https://cdn.oxfordnanoportal.com/software/analysis/dorado-0.2.1-linux-x64.tar.gz && \
tar zxf dorado-0.2.1-linux-x64.tar.gz && \
ln -s /dorado-0.2.1-linux-x64/bin/dorado /usr/local/bin/
# Install basecalling models
cd /dat
dorado download
# Convert FAST5 into POD5
cd /dat
mkdir pod5s
pod5 convert fast5 fast5s/*.fast5 pod5s --output-one-to-one fast5s
# Simplex calling
## Call canonical bases
dorado basecaller [email protected] pod5s/ | samtools view -Sh > reads.bam
## Call bases (including 5mCG and 5hmCG modifications)
dorado basecaller [email protected] pod5s/ --modified-bases 5mCG_5hmCG | samtools view -Sh > reads.bam
# Duplex calling
## Simplex call with --emit-moves
dorado basecaller [email protected] pod5s/ --emit-moves | samtools view -Sh > unmapped_reads_with_moves.bam
## Identify potential pairs
duplex_tools pair --output_dir ./pairs_from_bam unmapped_reads_with_moves.bam
## Stereo duplex basecall:
dorado duplex [email protected] pod5s/ --pairs pairs_from_bam/pair_ids_filtered.txt > reads_duplex.sam
gcloud compute instances create megalodon \
--machine-type a2-highgpu-2g \
--zone us-central1-a \
--boot-disk-size 2000GB \
--image-family cos-97-lts \
--image-project cos-cloud \
--maintenance-policy TERMINATE --restart-on-failure
gcloud compute ssh --zone us-central1-a megalodon
#toolbox gsutil -m cp -r gs:https://aryeelab-nanopore/medium_test /media/root/home/martin/
toolbox gsutil -m cp -r gs:https://aryeelab-nanopore/griffinlab/2022-06-24_hek293-hia5-k9me3 /media/root/home/martin/
# Install GPU drivers
sudo cos-extensions install gpu
sudo mount --bind /var/lib/nvidia /var/lib/nvidia
sudo mount -o remount,exec /var/lib/nvidia
# Configure artifact registry credentials
docker-credential-gcr configure-docker --registries us-central1-docker.pkg.dev
docker run --rm -it \
--volume /home/martin:/work \
--volume /var/lib/nvidia/lib64:/usr/local/nvidia/lib64 \
--volume /var/lib/nvidia/bin:/usr/local/nvidia/bin \
--device /dev/nvidia0:/dev/nvidia0 \
--device /dev/nvidia1:/dev/nvidia1 \
--device /dev/nvidia-uvm:/dev/nvidia-uvm \
--device /dev/nvidiactl:/dev/nvidiactl \
us-central1-docker.pkg.dev/aryeelab/docker/megalodon
# Download genome fasta
GENOME_URL="ftp:https://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank/Eukaryotes/vertebrates_mammals/Homo_sapiens/GRCh38/seqs_for_alignment_pipelines/GCA_000001405.15_GRCh38_full_analysis_set.fna.gz"
wget ${GENOME_URL} -P /work
# Obtain R9.4.1 modified base model from Rerio
git clone https://github.com/nanoporetech/rerio
rerio/download_model.py rerio/basecall_models/res_dna_r941_min_modbases-all-context_v001
# Call bases
SAMPLE="2022-06-24_hek293-hia5-k9me3"
mkdir -p /work/megalodon
time megalodon \
/work/${SAMPLE} \
--output-directory /work/megalodon/${SAMPLE} \
--guppy-server-path /usr/bin/guppy_basecall_server \
--guppy-params "-d /rerio/basecall_models/" \
--guppy-config res_dna_r941_min_modbases-all-context_v001.cfg \
--outputs basecalls mappings mod_mappings mods per_read_mods \
--reference /work/$(basename $GENOME_URL) --mod-motif Z CG 0 --mod-motif Y A 0 \
--devices cuda:all --processes 20
gcloud compute instances stop --zone us-central1-a megalodon
gcloud compute instances start --zone us-central1-a megalodon
# Performance on 2022-06-24_hek293-hia5-k9me3
VM GPUs processes Reads/s samples/s $/hour
a2-highgpu-1g 1 A100 5 21 1.74e+6 $3.67
a2-highgpu-1g 1 A100 10 37 3.1e+6 $3.67
a2-highgpu-1g 1 A100 20 37 3.04e+6 $3.67
a2-highgpu-2g 2 A100 20 67 5.13e+6 $7.35
a2-highgpu-4g 4 A100 10 69 5.63e+6 $14.69
a2-highgpu-4g 4 A100 20 111 9.01e+6 $14.69
a2-highgpu-4g 4 A100 40 171 1.38e+7 $14.69 # Drops to 106 reads/s due to full output queue
a2-highgpu-4g 4 A100 60 166 1.36e+7 $14.69
polaris 2 A100 10 67 5.2e+6
polaris 2 A100 20 119 9.37e+6
polaris 2 A100 30 164 1.25e+7 # Drops to 145 due to full output queue
polaris 1 A100 10 65 5.01e+6
polaris 1 A100 20 117 9.07e+6
polaris 1 A100 30 164 1.3e+7 # Drops to 145 due to full output queue
polaris 1 A100 40
polaris 1 A100 50
megalodon_extras aggregate run
a2-highgpu-2g
9:36:00
84954.53 per-read calls/s
# Get reference genome
GENOME_URL="ftp:https://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank/Eukaryotes/vertebrates_mammals/Homo_sapiens/GRCh38/seqs_for_alignment_pipelines/GCA_000001405.15_GRCh38_full_analysis_set.fna.gz"
wget ${GENOME_URL} -P /aryeelab/genomes
gunzip /aryeelab/genomes/$(basename $GENOME_URL)
# Obtain R9.4.1 modified base model from Rerio
cd /aryeelab/nanopore
git clone https://github.com/nanoporetech/rerio
rerio/download_model.py rerio/basecall_models/res_dna_r941_min_modbases-all-context_v001
gcloud auth configure-docker us-central1-docker.pkg.dev
singularity pull /aryeelab/singularity/megalodon.sif docker:https://us-central1-docker.pkg.dev/aryeelab/docker/megalodon
# Call bases
singularity shell --bind /aryeelab /aryeelab/singularity/megalodon.sif
SAMPLE_DIR="/aryeelab/nanopore/griffinlab/2022-06-24_hek293-hia5-k9me3"
OUT_DIR="/aryeelab/nanopore/griffinlab/megalodon/2022-06-24_hek293-hia5-k9me3_6mA_only"
#OUT_DIR="/tmp/2022-06-24_hek293-hia5-k9me3_6mA_only"
GENOME="/aryeelab/genomes/GCA_000001405.15_GRCh38_full_analysis_set.fna"
time megalodon \
${SAMPLE_DIR} \
--output-directory ${OUT_DIR} \
--guppy-server-path /usr/bin/guppy_basecall_server \
--guppy-params "-d /aryeelab/nanopore/rerio/basecall_models/" \
--guppy-config res_dna_r941_min_modbases-all-context_v001.cfg \
--outputs basecalls mappings mod_mappings mods per_read_mods \
--reference ${GENOME} --mod-motif Y A 0 \
--devices cuda:0 --processes 25
# --mod-motif Z CG 0
# Make bigwigs
CHROM_SIZES=/aryeelab/chrom_sizes/hg38.chrom.sizes
cp $CHROM_SIZES hg38.chrom.sizes
cd /aryeelab/nanopore/griffinlab/megalodon/2022-06-24_hek293-hia5-k9me3
#cat modified_bases.5mC.bed | cut -f 1,2,3,11 | sort -k1,1 -k2,2n > 5mC.percentage.bedgraph
#cat modified_bases.5mC.bed | cut -f 1,2,3,10 | sort -k1,1 -k2,2n > 5mC.coverage.bedgraph
time cat modified_bases.6mA.bed | cut -f 1,2,3,11 |sort -k1,1 -k2,2n > 6mA.percentage.bedgraph
time cat modified_bases.6mA.bed | cut -f 1,2,3,10 | sort -k1,1 -k2,2n > 6mA.coverage.bedgraph
#singularity run docker:https://4dndcic/4dn-bedgraphtobigwig bedGraphToBigWig 5mC.percentage.bedgraph hg38.chrom.sizes 5mC.percentage.bw
#singularity run docker:https://4dndcic/4dn-bedgraphtobigwig bedGraphToBigWig 5mC.coverage.bedgraph hg38.chrom.sizes 5mC.coverage.bw
time singularity run docker:https://4dndcic/4dn-bedgraphtobigwig bedGraphToBigWig 6mA.percentage.bedgraph hg38.chrom.sizes 6mA.percentage.bw
time singularity run docker:https://4dndcic/4dn-bedgraphtobigwig bedGraphToBigWig 6mA.coverage.bedgraph hg38.chrom.sizes 6mA.coverage.bw
Read Processing: 100%|█████████████████████████████████████████████████████████████████████████████████████| 1479118/1479118 [2:51:36<00:00, 143.65reads/s, samples/s=1.09e+7]
input queue capacity extract_signal : 0%| | 0/10000
output queue capacity basecalls : 0%| | 0/10000
output queue capacity mappings : 0%| | 0/10000
output queue capacity per_read_mods : 0%| | 1/10000
[23:48:40] Unsuccessful processing types:
9.8% ( 145645 reads) : No alignment
[23:48:40] Waiting for mods database to complete indexing
[01:32:17] Spawning modified base aggregation processes
[01:32:24] Aggregating 2941854010 per-read modified base statistics
[01:32:24] NOTE: If this step is very slow, ensure the output directory is located on a fast read disk (e.g. local SSD). Aggregation can be restarted using the `megalodon_extras aggregate run` command
Mods: 100%|████████████████████████████████████████████████████████████████████████████████████████████████| 2941854010/2941854010 [6:54:46<00:00, 118210.15 per-read calls/s]
[08:27:11] Mega Done
real 691m28.998s
user 7549m36.412s
sys 793m27.347s
(base) [martin@node01 2022-06-24_hek293-hia5-k9me3_gcp]$ time cat modified_bases.6mA.bed | cut -f 1,2,3,11 |sort -k1,1 -k2,2n > 6mA.percentage.bedgraph
real 441m47.441s
user 419m33.284s
sys 5m56.911s
time cat modified_bases.6mA.bed | cut -f 1,2,3,11 > tmp.cut
time sort -k1,1 -k2,2n tmp.cut > tmp.6mA.percentage.bedgraph
##########################
docker run --rm -it -v ${PWD}:/work ubuntu
apt-get update
apt-get -y install python3-pip cython3
pip install numpy
pip install megalodon
# --------
#pip install pyzmq
#pip install pyzmq --install-option="--zmq=bundled"
# apt-get -y install python3-zmq libzmq3-dev pkg-config
#apt-get -y install python3-dev python3-pip build-essential libzmq3-dev
pip install pyzmq
pip install --pre pyzmq
pip install pyzmq --install-option="--zmq=bundled"
# Verify GPU installation
sudo mount --bind /var/lib/nvidia /var/lib/nvidia
sudo mount -o remount,exec /var/lib/nvidia
/var/lib/nvidia/bin/nvidia-smi
# Test a docker container
docker run \
--volume /var/lib/nvidia/lib64:/usr/local/nvidia/lib64 \
--volume /var/lib/nvidia/bin:/usr/local/nvidia/bin \
--device /dev/nvidia0:/dev/nvidia0 \
--device /dev/nvidia-uvm:/dev/nvidia-uvm \
--device /dev/nvidiactl:/dev/nvidiactl \
gcr.io/google_containers/cuda-vector-add:v0.1
# CUDA images https://hub.docker.com/r/nvidia/cuda
docker run \
--volume /var/lib/nvidia/lib64:/usr/local/nvidia/lib64 \
--volume /var/lib/nvidia/bin:/usr/local/nvidia/bin \
--device /dev/nvidia0:/dev/nvidia0 \
--device /dev/nvidia-uvm:/dev/nvidia-uvm \
--device /dev/nvidiactl:/dev/nvidiactl \
nvidia/cuda:11.7.0-base-ubuntu20.04 nvidia-smi
# Interactive
See https://github.com/al-mcintyre/mcaller
#SAMPLE_NAME="20220107-ctcf-hia5"
WORK_DIR="${PWD}/work/20220107_CTCF_hia5_pilot/20220701_1749_MC-111988_0_FAR26597_5eec18a4"
GENOME_URL="ftp:https://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank/Eukaryotes/vertebrates_mammals/Homo_sapiens/GRCh38/seqs_for_alignment_pipelines/GCA_000001405.15_GRCh38_full_analysis_set.fna.gz"
curl ${GENOME_URL} -o ${WORK_DIR}/genome.fna.gz
cd ..
git clone --recursive https://github.com/aryeelab/nanopolish.git
cd nanopolish
docker build -t nanopolish .
cd ../nanopore-mods
cd Docker/minimap2
docker build -t minimap2 .
cd ../..
cd Docker/mcaller
docker build -t mcaller .
cd ../..
docker run -v ${WORK_DIR}/:/work/ --rm -it nanopolish /bin/bash
cd work
#zcat fastq_pass/*.fastq.gz > reads.fastq
zcat fastq_pass/*_0.fastq.gz > reads.fastq
seqkit stats reads.fastq > reads.fastq.stats
time /nanopolish/nanopolish index -d fast5_pass -s sequencing_summary*.txt reads.fastq 2> stderr.log
cat stderr.log | grep 'num reads' | sed 's/\[readdb\] //' | tr , '\n' | sed 's/^ //' > nanopolish-index.stats.txt
docker run -v ${WORK_DIR}/:/work/ --rm -it minimap2 /bin/bash
cd /work
time /minimap2/minimap2 -a genome.fna.gz reads.fastq | \
samtools view -Sb - | \
samtools sort -o reads.sorted.bam
samtools index reads.sorted.bam
samtools flagstat reads.sorted.bam > reads.sorted.bam.flagstat
docker run -v ${WORK_DIR}/:/work/ --rm -it nanopolish /bin/bash
cd /work
gunzip genome.fna.gz
/nanopolish/nanopolish eventalign -t 6 --scale-events -n -r reads.fastq -b reads.sorted.bam -g genome.fna 1> reads.eventalign.tsv 2> stderr.log
cat stderr.log | sed 's/\[post-run summary\] //' | tr , '\n' | sed 's/^ //' > eventalign.stats.txt
docker run -v ${WORK_DIR}/:/work/ --rm -it mcaller /bin/bash
cd /mCaller
gunzip genome.fna.gz
# 5 hours CTCF
time ./mCaller.py -m GATC -t 5 -r /work/genome.fna -d r95_twobase_model_NN_6_m6A.pkl -e /work/reads.eventalign.tsv -f /work/reads.fastq -b A
time ./make_bed.py -f /work/reads.eventalign.diffs.6 -d 1 -t 0
docker run -v ${WORK_DIR}/:/work/ --rm -it 4dndcic/4dn-bedgraphtobigwig /bin/bash
cd /work
CHROM_SIZES="hg38.chrom.sizes"
wget http:https://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/${CHROM_SIZES}
# A few reads are misassigned to the wrong alt contig? Temp fix:
bedClip -verbose=2 reads.methylation.summary.bed ${CHROM_SIZES} reads.methylation.summary.clipped.bed
cat reads.methylation.summary.clipped.bed | cut -f1,2,3,5 | sort -k1,1 -k2,2n > m6a.ratio.bedgraph
cat reads.methylation.summary.clipped.bed | cut -f1,2,3,7 | sort -k1,1 -k2,2n > m6a.coverage.bedgraph
bedGraphToBigWig m6a.ratio.bedgraph ${CHROM_SIZES} m6a.ratio.bw
bedGraphToBigWig m6a.coverage.bedgraph ${CHROM_SIZES} m6a.coverage.bw
cromwell run -i test_data/test-run-1.cpu.json preprocess_flowcell.wdl
You can include the path to a custom Cromwell config file by something like:
JAVA_OPTS="-Dconfig.file=/Users/maryee/cromwell/cromwell.conf"
cromwell run -i test_data/test-run-1.json preprocess_flowcell.wdl
(See https://cromwell.readthedocs.io/en/latest/tutorials/ConfigurationFiles/)
This can be used to specify a cache database, for example:
docker run -p 3306:3306 --name cromwell_mysql -e MYSQL_ROOT_PASSWORD=pass123 -e MYSQL_DATABASE=cromwell -e MYSQL_USER=cromwell -e MYSQL_PASSWORD=pass123 -d mysql/mysql-server:5.5
(See https://cromwell.readthedocs.io/en/latest/tutorials/PersistentServer/)
e.g.
docker run --rm -it aryeelab/guppy
java -jar -Dconfig.file=lsf.bash.conf /data/molpath/software/cromwell-39.jar run preprocess_flowcell_singularity.wdl -i test_data/test-run-1.json
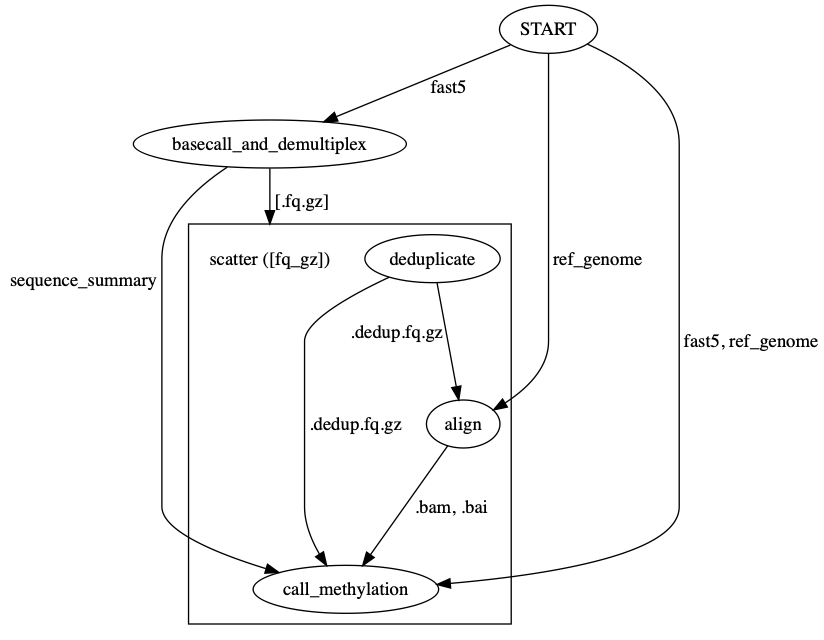
You can use womtool (part of cromwell) to output a workflow graph in .dot format:
womtool graph preprocess_flowcell.wdl > preprocess_flowcell.dot
This graph can be edited if necessary (such as to label edges with inputs/outputs), and then visualized with graphviz (brew install graphviz):
dot preprocess_flowcell.dot -Tpng -o preprocess_flowcell.png
The workflow graph below is produced in this way.