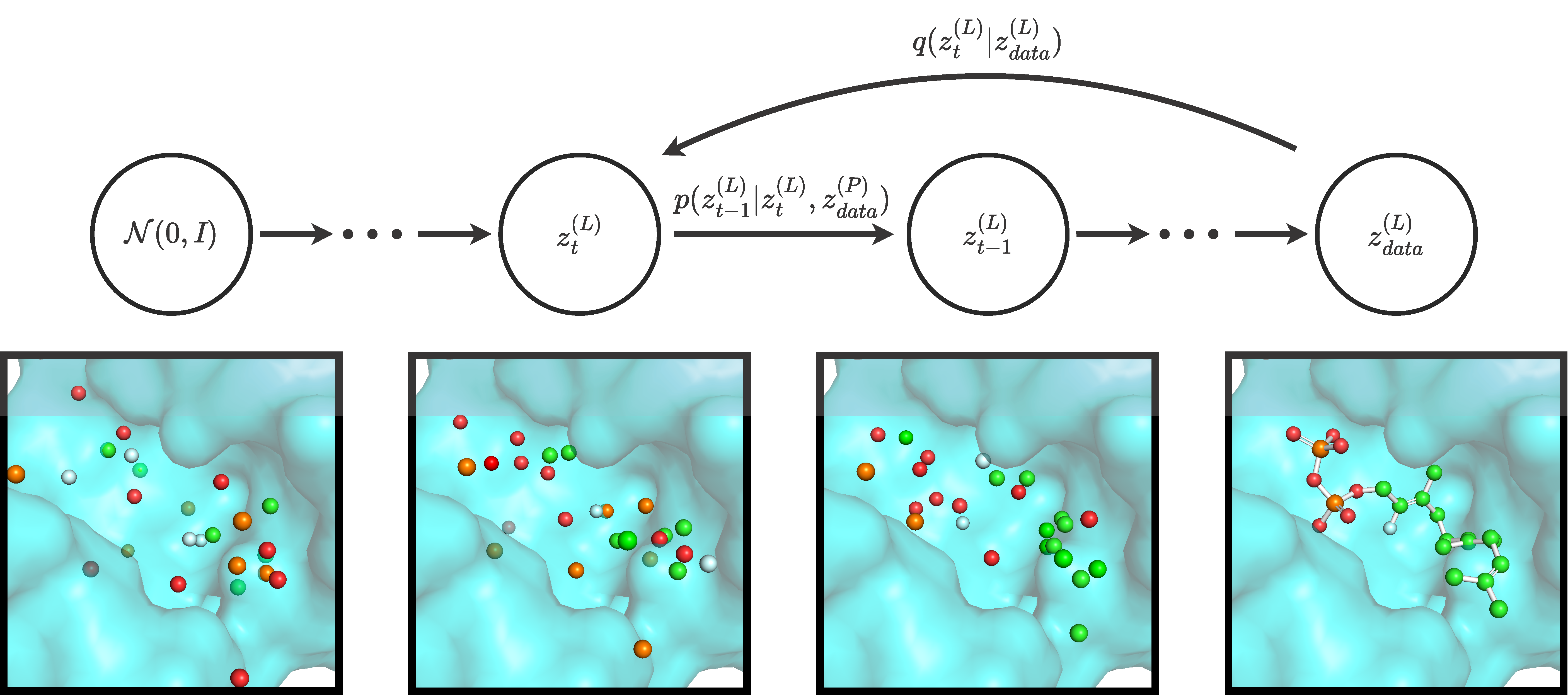
Official implementation of DiffSBDD, an equivariant diffusion model for structure-based drug design, by Arne Schneuing, Charles Harris, Yuanqi Du, Kieran Didi, Arian Jamasb, Ilia Igashov, Weitao Du, Carla Gomes, Tom Blundell, Pietro Lio, Max Welling, Michael Bronstein & Bruno Correia.
conda create -n sbdd-env
conda activate sbdd-env
conda install pytorch cudatoolkit=10.2 -c pytorch
conda install -c conda-forge pytorch-lightning
conda install -c conda-forge wandb
conda install -c conda-forge rdkit
conda install -c conda-forge biopython
conda install -c conda-forge imageio
conda install -c anaconda scipy
conda install -c pyg pytorch-scatter
conda install -c conda-forge openbabel
conda install seabornThe code was tested with the following versions
| Software | Version |
|---|---|
| Python | 3.10.4 |
| CUDA | 10.2.89 |
| PyTorch | 1.12.1 |
| PyTorch Lightning | 1.7.4 |
| WandB | 0.13.1 |
| RDKit | 2022.03.2 |
| BioPython | 1.79 |
| imageio | 2.21.2 |
| SciPy | 1.7.3 |
| PyTorch Scatter | 2.0.9 |
| OpenBabel | 3.1.1 |
Pre-trained models can be downloaded from Zenodo.
- CrossDocked, conditional
$C_\alpha$ model - CrossDocked, joint
$C_\alpha$ model - CrossDocked, conditional full-atom model
- CrossDocked, joint full-atom model
- Binding MOAD, conditional
$C_\alpha$ model - Binding MOAD, joint
$C_\alpha$ model - Binding MOAD, conditional full-atom model
- Binding MOAD, joint full-atom model
These simple step-by-step examples provide an easy entry point to generating molecules with DiffSBDD. More details about training and sampling scripts are provided below.
Before we run the sampling scripts we need to download a model checkpoint:
wget -P checkpoints/ https://zenodo.org/record/8183747/files/crossdocked_fullatom_cond.ckptIt will be stored in the ./checkpoints folder.
Using the trained model weights, we can sample new ligands with a single command. In this example, we use the protein with PDB ID 3RFM that can be found in the example folder.
The PDB file contains a reference ligand in chain A at residue number 330 that we can use to specify the designated binding pocket.
The following command will generate 20 samples and save them in a file called 3rfm_mol.sdf in the ./example folder.
python generate_ligands.py checkpoints/crossdocked_fullatom_cond.ckpt --pdbfile example/3rfm.pdb --outfile example/3rfm_mol.sdf --ref_ligand A:330 --n_samples 20Instead of specifying the chain and residue number we can also provide an SDF file with the reference ligand:
python generate_ligands.py checkpoints/crossdocked_fullatom_cond.ckpt --pdbfile example/3rfm.pdb --outfile example/3rfm_mol.sdf --ref_ligand example/3rfm_B_CFF.sdf --n_samples 20If no reference ligand is known, the binding pocket can also be specified as a list of residues as described below.
To design molecules around fixed substructures (scaffold elaboration, fragment linking etc.) you can run the inpaint.py script.
Here, we demonstrate its usage with a fragment linking example. Similar to generate_ligands.py, the inpainting script allows us to define pockets based on a reference ligand in SDF format
or with a chain and residue identifier (if it is in the PDB).
The easiest way to fix substructures is to provide them in a separate SDF file using the --fix_atoms flag.
However, the script also accepts a list of atom names which must correspond to the atoms of the reference ligand in the PDB file, e.g. --fix_atoms C1 N6 C5 C12.
python inpaint.py checkpoints/crossdocked_fullatom_cond.ckpt --pdbfile example/5ndu.pdb --outfile example/5ndu_linked_mols.sdf --ref_ligand example/5ndu_C_8V2.sdf --fix_atoms example/fragments.sdf --center ligand --add_n_nodes 10Note that the --center ligand option tells DiffSBDD to sample the additional atoms near the center of mass of the fixed substructure, which is not always ideal or desired.
For instance, the inputs could be two fragments with very different sizes, in which case the random noise will be sampled very close to the larger fragment.
We currently also support sampling in the pocket center (--center pocket) but in some cases neither of these two options might be suitable and a problem-specific solution is warranted to avoid bad results.
Another important parameter is --add_n_nodes which determines how many new atoms will be added. If it is not provided, a random number will be sampled.
You can use DiffSBDD to optimize existing molecules for given properties via the optimize.py script.
python optimize.py checkpoints/crossdocked_fullatom_cond.ckpt --pdbfile example/5ndu.pdb --outfile output.sdf --ref_ligand example/5ndu_C_8V2.sdf --objective sa --population_size 100 --evolution_steps 10 --top_k 10 --timesteps 100Important parameters in the evolutionary algorithum are:
--objective: The optimization objective. Currently supports 'qed' for Quantitative Estimate of Drug-likeness and 'sa' for Synthetic Accessibility. Custom objectives can be implemented within the code.--population_size: The size of the molecule population to maintain across the optimization generations.--evolution_steps: The number of evolutionary steps (generations) to perform during the optimization process.--top_k: The number of top-scoring molecules to select from one generation to the next.--timesteps: The number of noise-denoise steps to use in the optimization algorithum. Defaults to 100 (out of T=500).
Download and extract the dataset as described by the authors of Pocket2Mol: https://github.com/pengxingang/Pocket2Mol/tree/main/data
Process the raw data using
python process_crossdock.py <crossdocked_dir> --no_HDownload the dataset
wget https://www.bindingmoad.org/files/biou/every_part_a.zip
wget https://www.bindingmoad.org/files/biou/every_part_b.zip
wget https://www.bindingmoad.org/files/csv/every.csv
unzip every_part_a.zip
unzip every_part_b.zipProcess the raw data using
python -W ignore process_bindingmoad.py <bindingmoad_dir>Add the --ca_only flag to create a dataset with
Sampled molecules can be found on Zenodo.
Starting a new training run:
python -u train.py --config <config>.ymlResuming a previous run:
python -u train.py --config <config>.yml --resume <checkpoint>.ckptTo sample small molecules for a given pocket with a trained model use the following command:
python generate_ligands.py <checkpoint>.ckpt --pdbfile <pdb_file>.pdb --outfile <output_file> --resi_list <list_of_pocket_residue_ids>For example:
python generate_ligands.py last.ckpt --pdbfile 1abc.pdb --outfile results/1abc_mols.sdf --resi_list A:1 A:2 A:3 A:4 A:5 A:6 A:7 Alternatively, the binding pocket can also be specified based on a reference ligand in the same PDB file:
python generate_ligands.py <checkpoint>.ckpt --pdbfile <pdb_file>.pdb --outfile <output_file> --ref_ligand <chain>:<resi>or with a separate SDF file:
python generate_ligands.py <checkpoint>.ckpt --pdbfile <pdb_file>.pdb --outfile <output_file> --ref_ligand <ref_ligand>.sdfOptional flags:
| Flag | Description |
|---|---|
--n_samples |
Number of sampled molecules |
--num_nodes_lig |
Size of sampled molecules |
--timesteps |
Number of denoising steps for inference |
--all_frags |
Keep all disconnected fragments |
--sanitize |
Sanitize molecules (invalid molecules will be removed if this flag is present) |
--relax |
Relax generated structure in force field (does not consider the protein and might introduce clashes) |
--resamplings |
Inpainting parameter (doesn't apply if conditional model is used) |
--jump_length |
Inpainting parameter (doesn't apply if conditional model is used) |
test.py can be used to sample molecules for the entire testing set:
python test.py <checkpoint>.ckpt --test_dir <bindingmoad_dir>/processed_noH/test/ --outdir <output_dir> --sanitizeThere are different ways to determine the size of sampled molecules.
--fix_n_nodes: generates ligands with the same number of nodes as the reference molecule--n_nodes_bias <int>: samples the number of nodes randomly and adds this bias--n_nodes_min <int>: samples the number of nodes randomly but clamps it at this value
Other optional flags are analogous to generate_ligands.py.
inpaint.py can be used for partial ligand redesign with the conditionally trained model, e.g.:
python inpaint.py <checkpoint>.ckpt --pdbfile <pdb_file>.pdb --outfile <output_file> --ref_ligand <chain>:<resi> --fix_atoms C1 N6 C5 C12--add_n_nodes controls the number of newly generated nodes. Other options are the same as before.
For assessing basic molecular properties create an instance of the MoleculeProperties class and run its evaluate method:
from analysis.metrics import MoleculeProperties
mol_metrics = MoleculeProperties()
all_qed, all_sa, all_logp, all_lipinski, per_pocket_diversity = \
mol_metrics.evaluate(pocket_mols)evaluate() expects a list of lists where the inner list contains all RDKit molecules generated for one pocket.
@article{schneuing2022structure,
title={Structure-based Drug Design with Equivariant Diffusion Models},
author={Schneuing, Arne and Du, Yuanqi and Harris, Charles and Jamasb, Arian and Igashov, Ilia and Du, Weitao and Blundell, Tom and Li{\'o}, Pietro and Gomes, Carla and Welling, Max and Bronstein, Michael and Correia, Bruno},
journal={arXiv preprint arXiv:2210.13695},
year={2022}
}