Wrath is a program for the visualisation and identification of candidate structural variants (SVs) from linked read data. Wrath calculates barcode sharing between windows of a given chromosome and produces heatmap plots that are useful to explore the data and identify candidate SVs. The pipeline can also automatically detect large SVs by using a double approach of z-scores threasholds and prediction bands from modelling of the data.
- Running Wrath for Structural Variant detection
- Requirements
- Input files
- Output
- Test example run
- Running Wrath on multiple chromosomes
- Citing Wrath
The main script wrath runs the main version of Wrath. The pipeline calculates barcode sharing between windows of a chosen size in a given chromosome and produces heatmap plots of barcode sharing. The main pipeline can also automatically detect large SVs. Default window size is 50kbp.
To run this scripts barcodes need to be encoded in the BX tag beforehand. To do that you can use the barcode_parsing utility.
A typical command looks like:
wrath -g reference_genome.fa -c chromosome_name -w 50000 -a list_of_bam_files.txt -t 15
Input options are:
Wrath: wrapped analysis of tagged haplotypes
DESCRIPTION:
Program produces a jaccard matrix camparing the barcode content between all pairs windows whithin a chromosome.
wrath [-h] [-g FASTAFILE] [-c CHROMOSOMENAME] [-w WINDOWSIZE] [-a FILELIST] [-t THREADS] [-p] [-v] [-x STEP] [-l] [-s START] [-e END]
OPTIONS:
-h show this help text
-g FASTAFILE reference genome
-c CHROMOSOMENAME chromosome
-w WINDOWSIZE window size
-a FILELIST list of bam files with paths of the individuals of the population/phenotype of interest
-t THREADS threads to use
-p skip plotting the heatmap
-x STEP start from a given step. Note that this only works if filenames match those expected by wrath. Possible step options are: makewindows, getbarcodes, matrix, outliers (only if -l given) or plot
-l automatic detection of SVs
-v verbose (only for the matrix generating step)
-s START start position to subset windows
-e END end position to subset windows
Versions shown are the ones used for development. Other versions might work but have not been tested.
Command line programs:
Python (version 3.6 or higher):
pip install -U numpy seaborn matplotlib pandas scikit-learn pysamR (version 4.0.3 or higher):
In R, run
# The easiest way to get ggplot2, tidyr and dplyr is to install the whole tidyverse:
install.packages("tidyverse")
#and then nlraa
install.packages("nlraa")The input necessary is a reference genome in fasta format and a list of the sample bam files that need to be analysed including their paths.
Like:
/home/samples/bams/group1/sample1.bam
/home/samples/bams/group1/sample2.bam
/home/samples/bams/group2/sample3.bam
/home/samples/bams/group2/sample4.bam
Wrath will create a directory with this structure (when running with default options):
wrath_out/
├── beds
├── matrices
├── outliers
├── plots
└── SVs
Table of automatically detected SVs for a given chromosome (in csv format). Columns include: SV id, chromosome name, start position, end position and SV length.
SV_id,chromsome,start,end,length
5,Herato1910,10000,3170000,3160000
15,Herato1910,540000,2160000,1620000
13,Herato1910,260000,1110000,850000
19,Herato1910,90000,620000,530000
16,Herato1910,1600000,2130000,530000
6,Herato1910,2530000,2940000,410000
12,Herato1910,940000,1090000,150000
2,Herato1910,600000,750000,150000
0,Herato1910,4820000,4910000,90000
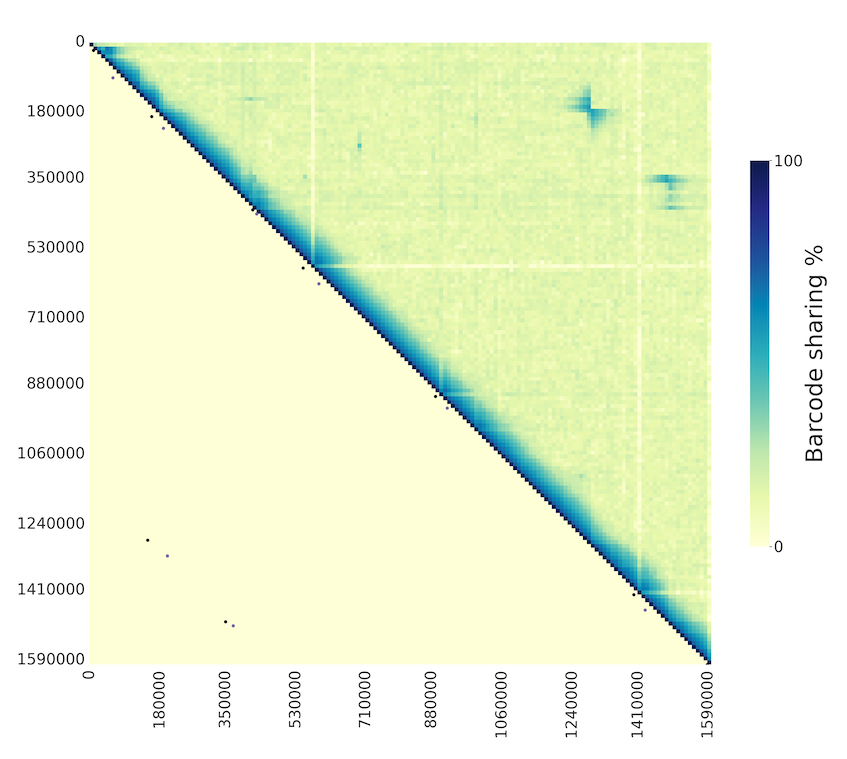
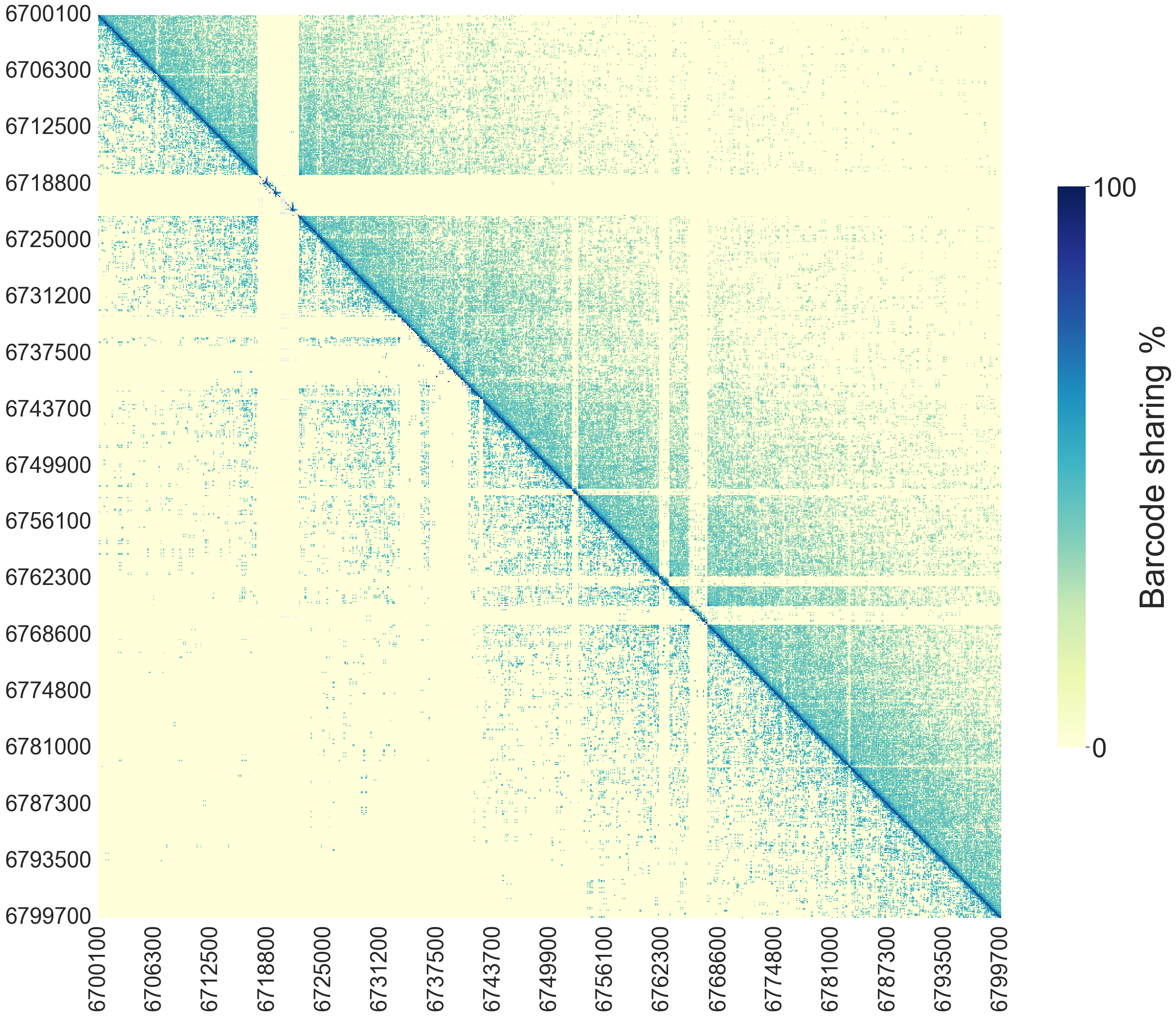
Heatmap plot of barcode sharing between windows of a given chromosome. If automatic detection of SVs is used, the lower triangle of the plot will indicate the points where SVs have been detected, while the upper triangle will show barcode sharing between windows.
If automatic detection of SVs is not enabled, the upper triangle will show barcode sharing between windows and lower triangle will be empty.
If comparing two populations that differ in a structural variant, it can be useful to plot the barcode sharing between windows of the two populations. This can be done by using the script sv_detection/plot_2matrices_tegether.py and providing a list of bam files for each population. The plot will show the barcode sharing between windows of each population, one in the upper triangle and the other in the lower triangle.
-
Window Beds: To calculate barcode sharing between windows, first Wrath splits the chromosome into n windows of size m. The coordinates of those windows are stored in a bed file in the directory beds.
-
Barcode Beds: Barcodes are extracted from the bam files, and their leftmost mapping position is stored in a gzipped bed file in beds. Tabix indexes are also created.
-
Matrices: Barcode sharing between pairs of windows is calculated and stored in an identity matrix of nxn dimensions. A Jaccard index is calculated for each pair of windows:
Where J is the Jaccard distance, and A and B are windows 1 and 2, respectively.
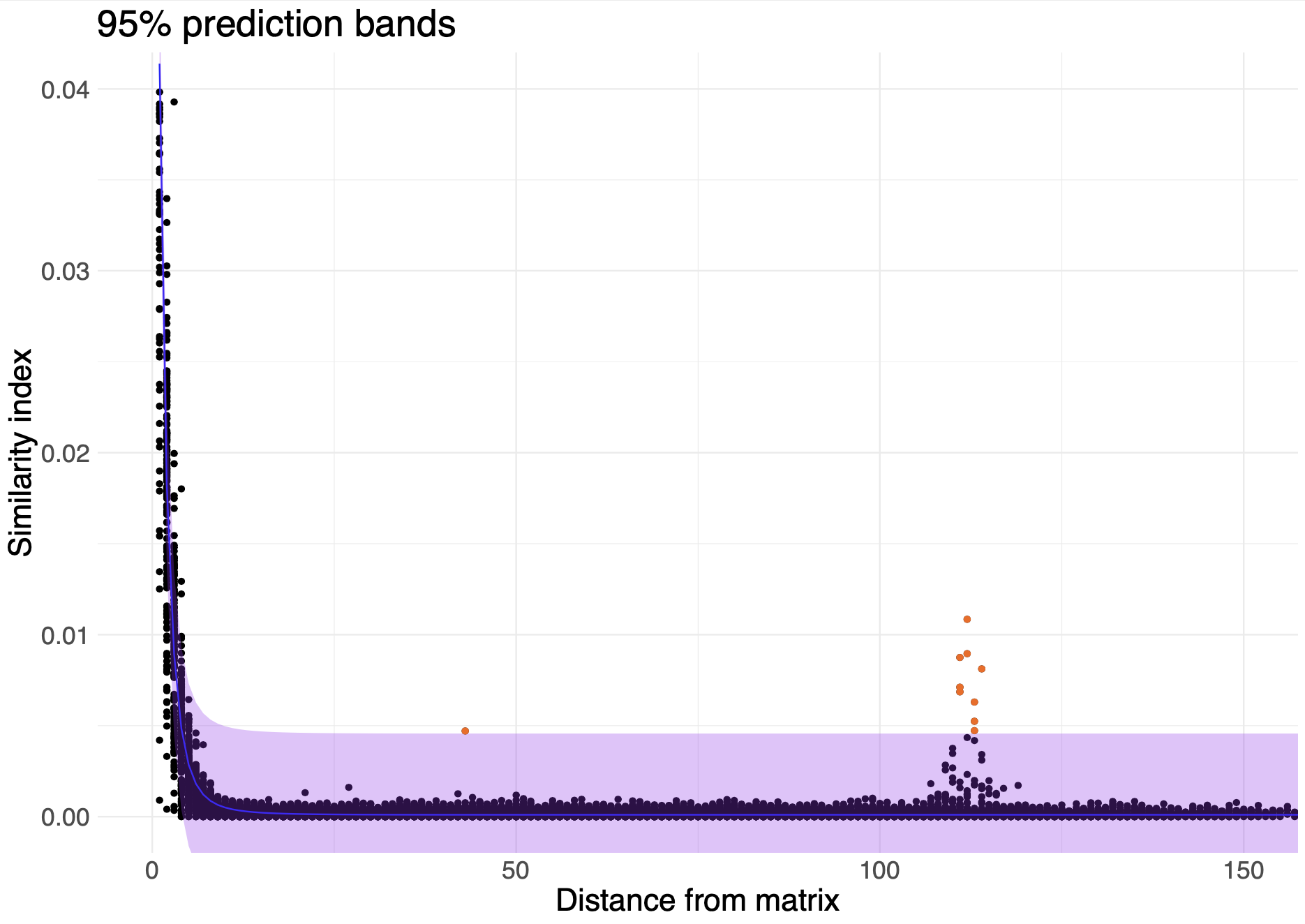
- Outliers: We calculate and store the distance of each comparison to the diagonal. Then, using this distance and the Jaccard index value of the comparison, we calculate z scores and, separately, we fit a double exponential decay model, such that:
The model is fit, and 95% prediction bands are calculated from it such that:
Any points outside the z score threshold (2 by default) and above or below the prediction bands are defined as outliers and stored in the outliers directory. Z score threashold and predition bands can be changed in the script outliers.R.
Several values are stored for each outlier: row number, column number, jaccard distance, y estimate of the model, estimated error, 2.5 quantile, 97.5 quantile and a definition of whether it is an 'upper' or 'lower' outlier.
To test Wrath's installation and prerequisites, there is a complete example test set in example_run/complete_heliconius_example. Follow the README to run the example.
The easiest way to run Wrath on multiple chromosomes is to run in parallely. If running on a cluster and using a shceduling system such as SLURM, an array can be used to run a job for each chromosome. An example is found in example array.
If you use Wrath please cite the our MBE paper:
Anna Orteu, Marek Kucka, Ian J Gordon, Ivy Ng’iru, Eva S M van der Heijden, Gerard Talavera, Ian A Warren, Steve Collins, Richard H ffrench-Constant, Dino J Martins, Yingguang Frank Chan, Chris D Jiggins, Simon H Martin, Transposable Element Insertions Are Associated with Batesian Mimicry in the Pantropical Butterfly Hypolimnas misippus, Molecular Biology and Evolution, Volume 41, Issue 3, March 2024, msae041, https://doi.org/10.1093/molbev/msae041