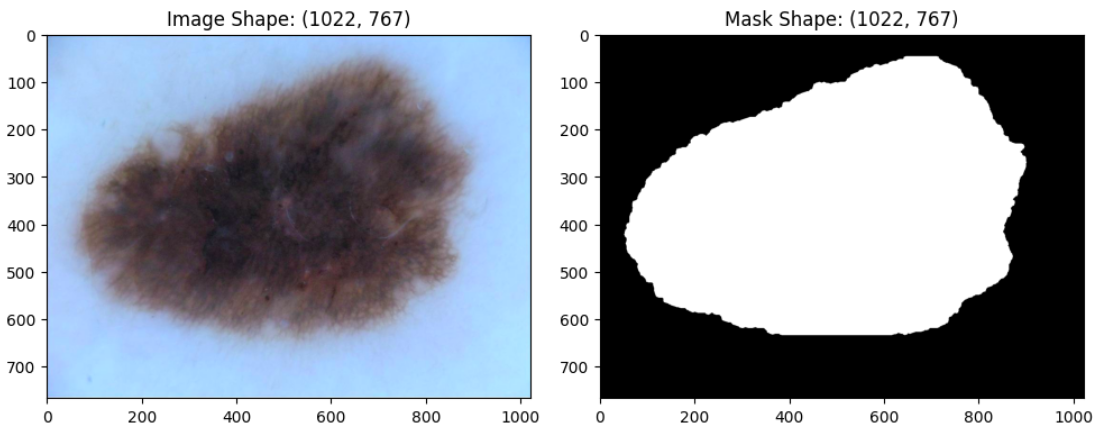
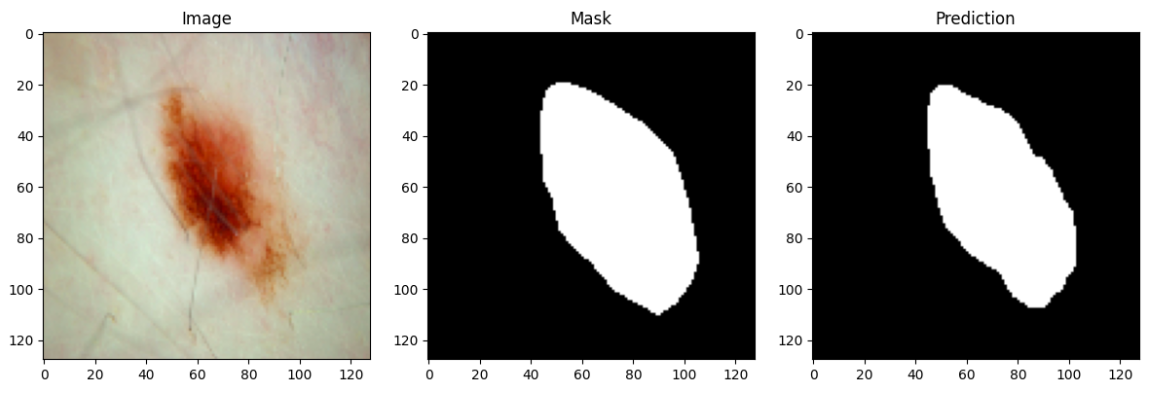
The primary objective of this challenge is to develop image analysis tools to enable the automated diagnosis of melanoma from dermoscopic images.
Skin cancer poses a significant public health concern. Melanoma, the most lethal form of skin cancer, accounts for the majority of skin cancer-related fatalities. Despite its high mortality rate, early detection of melanoma can lead to a survival rate exceeding 95%. Therefore, the objective is to achieve the highest possible performance of melanoma detecting model.
The training dataset contains 900 dermoscopic lesion images in JPEG format, along with the corresponding segmentation mask.
The ISIC Lesion segmentation dataset is originally introduced in the ISIC Challenge webpage.
- A U-Net model (implemented from scratch in PyTorch) with a reduced number of parameters compared to the vanilla version.
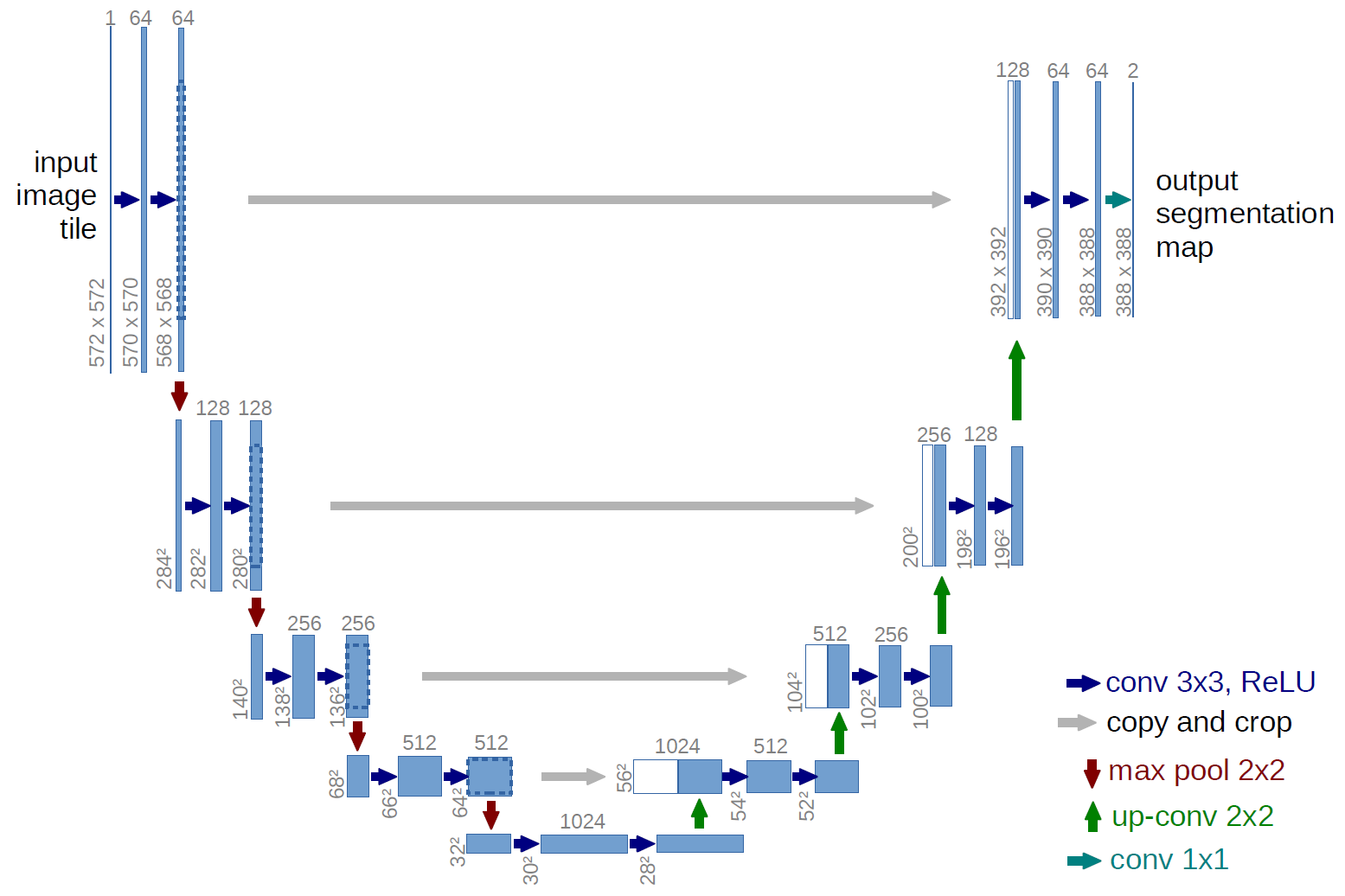
- The file named
segmentation_model.pthcontains the weight parameters of the trained model. - Execute the
isic-2016-challenge-lesion-segmentation.ipynbnotebook to train the designated model.
- Sensitivity score: 0.871
- Specificity score: 0.969
- Pixel accuracy: 0.957
- Jaccard score: 0.805
- Dice score: 0.885