BioTorch Provides:
🧠 Implementations of layers, models and biologically-motivated learning algorithms. It allows to load existing state-of-the-art models, easy creation of custom models and automatic conversion of existing models.
🧠 A framework to train, evaluate and benchmark different biologically plausible learning algorithms in a selection of datasets. It is focused on the principles of PyTorch design and research reproducibility. Configuration files that include the choice of a fixed seed and deterministic math and CUDA operations are provided.
🧠 A place of colaboration, ideas sharing and discussion.
| Name | Mode | Official Implementations |
|---|---|---|
| Feedback Alignment | 'fa' |
N/A |
| Direct Feedback Alignment | 'dfa' |
[Torch] |
| Sign Symmetry | ['usf', 'brsf', 'frsf'] |
[PyTorch] |
| Layer Weight Alignment | Layer Weight Norm Ratio |
|---|---|
 |
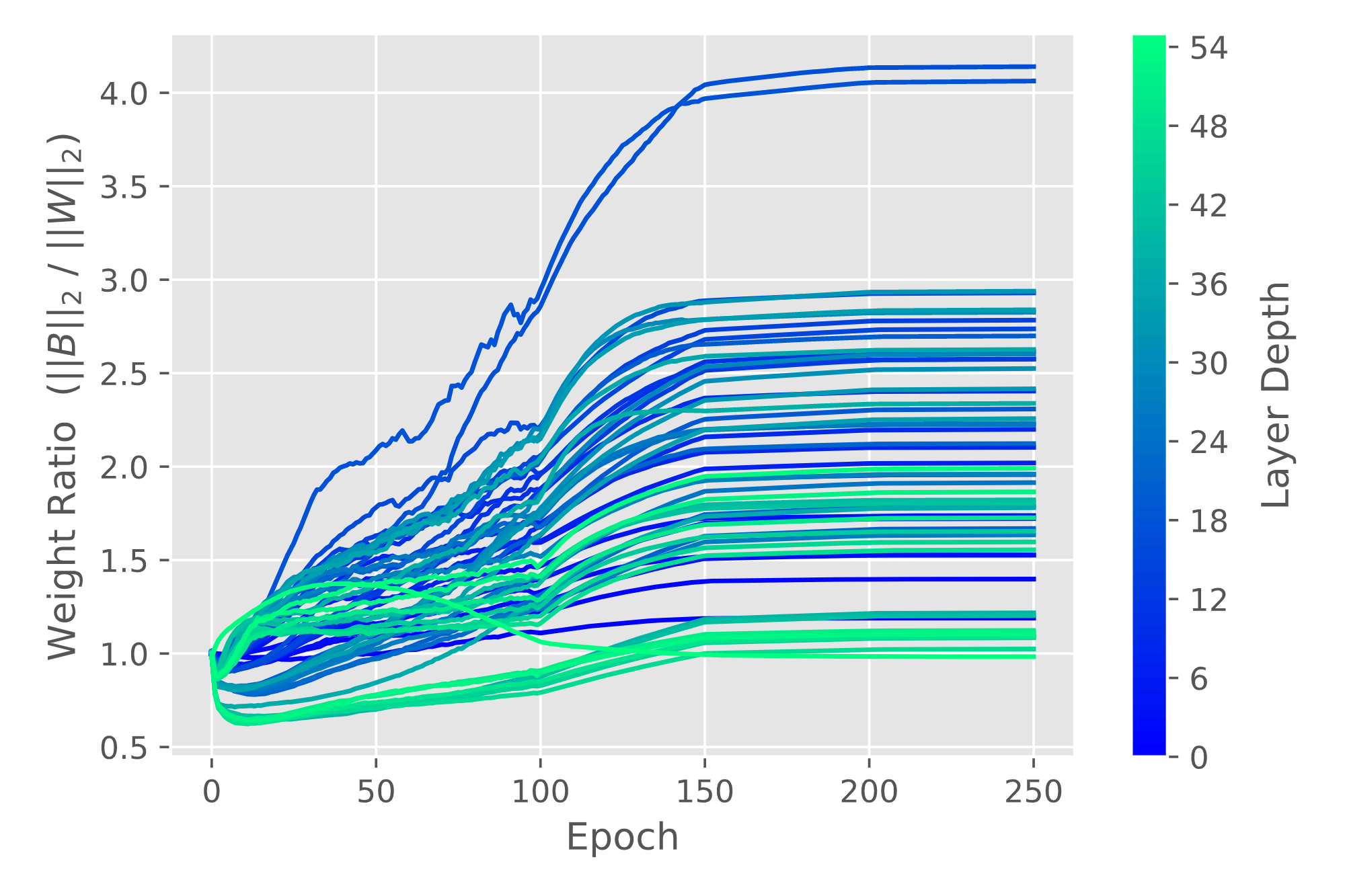 |
from biotorch.models.fa import resnet18
model = resnet18()
import torch.nn.functional as F
from biotorch.layers.usf import Conv2d, Linear
class Model(nn.Module):
def __init__(self):
super(Model, self).__init__()
self.conv1 = Conv2d(in_channels=64, out_channels=128, kernel_size=3)
self.fc = Linear(in_features=256, out_features=10)
def forward(self, x):
out = F.relu(self.conv1(x))
out = F.avg_pool2d(out, out.size()[3])
return self.fc(out)
model = Model()
from torchvision.models import alexnet
from biotorch.module.biomodule import BioModule
model = BioModule(module=alexnet(), mode='frsf')
python benchmark.py --config benchmark_configs/mnist/fa.yaml
If you want the experiment to be reproducible, check that you have specified a seed and the parameter deterministicis set to true in the configuration file yaml. That will apply all the PyTorch reproducibility steps.
If you are running your experiment on GPU add the extra environment variable CUBLAS_WORKSPACE_CONFIG.
CUBLAS_WORKSPACE_CONFIG=:4096:8 python benchmark.py --config benchmark_configs/mnist/fa.yaml
Click here to learn more about the configuration file API.
We are hosted in PyPI, you can install the library using pip:
pip install biotorch
Or from source:
git clone https://github.com/jsalbert/biotorch.git
cd biotorch
script/setup
If you want to contribute to the project please read the CONTRIBUTING section. If you found any bug please don't hesitate to comment in the Issues section.
Backpropagation is the default algorithm for training deep neural networks due to its simplicity, efficiency and high convergence rate. However, its requirements make it impossible to be implemented in a human brain. In recent years, more biologically plausible learning methods have been proposed. Some of these methods can match backpropagation accuracy, and simultaneously provide other extra benefits such as faster training on specialized hardware (e.g., ASICs) or higher robustness against adversarial attacks. While the interest in the field is growing, there is a necessity for open-source libraries and toolkits to foster research and benchmark algorithms. In this paper, we present BioTorch, a software framework to create, train, and benchmark biologically motivated neural networks. In addition, we investigate the performance of several feedback alignment methods proposed in the literature, thereby unveiling the importance of the forward and backward weight initialization and optimizer choice. Finally, we provide a novel robustness study of these methods against state-of-the-art white and black-box adversarial attacks.
Preprint here, feedback welcome!
Contact: [email protected]
If you use our code in your research, you can cite our paper:
@misc{sanfiz2021benchmarking,
title={Benchmarking the Accuracy and Robustness of Feedback Alignment Algorithms},
author={Albert Jiménez Sanfiz and Mohamed Akrout},
year={2021},
eprint={2108.13446},
archivePrefix={arXiv},
primaryClass={cs.LG}
}
