- About Nerpa
1.1 Nerpa pipeline
1.2 Supported data types - Installation
2.1. Installation via conda
2.2. Installation from source code - Running Nerpa
3.1. Quick start
3.2. Command-line options
3.3. Output files
3.4. Use case 1: inspecting a genome
3.5. Use case 2: inspecting a compound
3.6. Use case 3: inspecting both - Citation
- Feedback and bug reports
Nerpa is a tool for linking biosynthetic gene clusters (BGCs) to known nonribosomal peptides (NRPs). The paper revealing details of the Nerpa algorithm and demonstrating the practical application of the tool is freely available here. The software is developed in the Center for Algorithmic Biotechnology, St. Petersburg State University; the project page is here.
This manual will help you to install and run Nerpa. Nerpa version 1.0.0 was released on September 13, 2021. The tool is dual-licensed and is available under GPLv3 or Creative Commons BY-NC-SA 4.0, see LICENSE.txt for details.
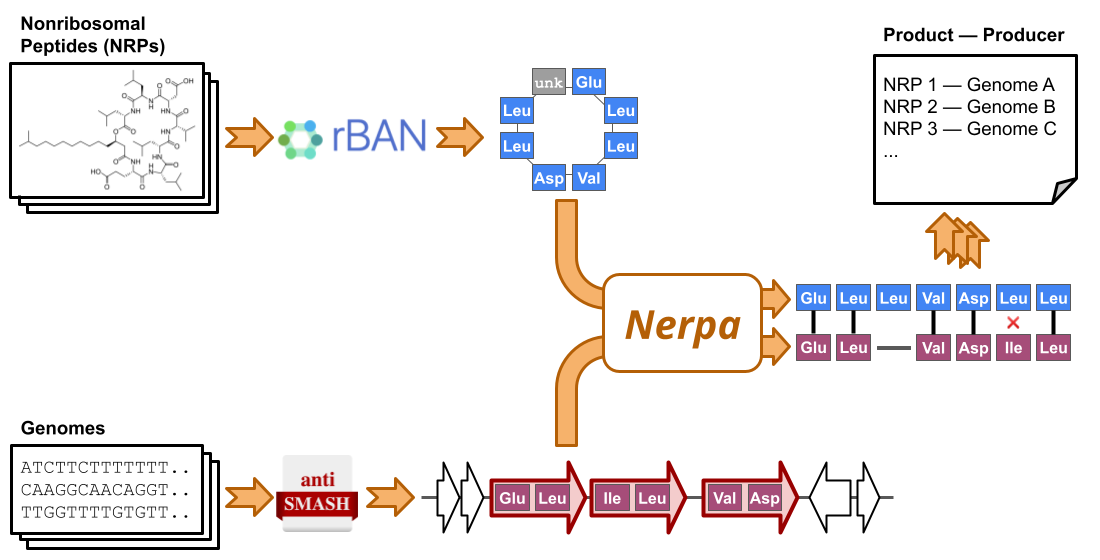
The simplified Nerpa pipeline is depicted in the figure below.
Nerpa takes as input an NRP database and genome sequences. The pipeline starts by detecting (i) linear representations of the database structures (using rBAN), and (ii) tentative NRP synthetase assembly lines along with respective sequences of genome-predicted residues (using antiSMASH). Afterward, Nerpa (iii) aligns the retrieved sequences against each other in an all-vs-all manner (using the Needleman–Wunsch algorithm and the Nerpa scoring described in the paper). Finally, Nerpa (iv) reports best matches per genome, per NRP, and combined.
For genome sequences:
- Recommended: antiSMASH output JSON format; e.g., downloaded from the antiSMASH web server after processing your raw genome sequence.
- Also accepted: raw genome sequences in the FASTA and GenBank formats; in this case Nerpa will predict NRP BGCs in them with antiSMASH (should be installed separately and present in
PATHor provided to Nerpa via--antismash-path).
For NRP structures:
- Recommended: isomeric SMILES format; Nerpa distinguishes between L- and D-configurations of amino acids, so the use of the isomeric format leads to more accurate results.
- Also accepted: any other SMILES, i.e., without stereochemistry information.
- Also accepted (for advanced users): custom NRP monomer graph format, check out this file as a represenative example, the list of accepted monomers is here, use
X0for anything else.
Note: you can use free online converters to get (isomeric) SMILES from other popular chemical formats such as MDL MOL or INCHI, e.g., this one from UNM. Alternatively, there are many command-line convertors, e.g. molconvert, or programming libraries, e.g. RDKit.
You can install Nerpa using one of the two available options described below. In either case, the Nerpa package includes rBAN but lacks antiSMASH. So, if you plan to use Nerpa with raw genome sequences (FASTA or GenBank) rather than antiSMASH-processed files (JSON), you need also to install antiSMASH on your computer.
This is the easiest and the most recommended way since conda will automatically manage all dependencies. To install Nerpa, just type
conda install -c bioconda nerpa
If you are not familiar with conda/bioconda, please consult with their documentation.
Before compiling Nerpa from the source code, please satisfy the following requirements:
- 64-bit Linux system or macOS
- g++ v.5.2+ or clang v.3.2+
- cmake v.3.5+
For running Nerpa you would also need:
- Python v.3.6+
- Python libraries RDKit and networkx
- java (for running rBAN, i.e., only needed if you specify NRPs in the SMILES format)
First, get the source code of the release
wget https://github.com/ablab/nerpa/releases/download/v1.0.0/nerpa-1.0.0.tar.gz
tar xzf nerpa-1.0.0.tar.gz
cd nerpa-1.0.0
or try the cutting-edge development code from GitHub
git clone https://github.com/ablab/nerpa.git
cd nerpa
Next, build Nerpa in ./bin inside the current directory by typing
./install.sh
or install the tool into another directory by running
PREFIX=<destination_dir> ./install.sh
For example,
PREFIX=/usr/local ./install.sh
will install Nerpa into /usr/local/bin/.
Note: you should use an absolute path for <destination_dir>.
If the installation was successful, you should see the NRPsMatcher and nerpa.py executable files in the ./bin directory (or <destination_dir>/bin if you specified PREFIX).
We recommend adding the Nerpa bin directory to PATH. In this case, you can run Nerpa simply as nerpa.py from anywhere; otherwise, you would need to specify path from the current directory to ./nerpa.py. All running examples below assume that Nerpa is in PATH.
To test your installation, first, try to get the list of the Nerpa command-line options:
nerpa.py -h
Then, try any example from the Quick start section and ensure the log contains no error messages.
If you have any problems, please don't hesitate to contact us.
Download test data provided with the release (also available in the GitHub repository in test_data):
wget https://github.com/ablab/nerpa/releases/download/v1.0.0/test_data.tar.gz
tar xzf test_data.tar.gz
Example 1: three reference genomes downloaded from NCBI RefSeq and pre-processed with antiSMASH v.3 against three NRP structures pre-processed with Nerpa (custom monomer graph format)
nerpa.py -a test_data/NCBI_subset/genome_predictions/ --structures test_data/NCBI_subset/structure.info.monomers
Example 2: five BGCs sequences downloaded from MIBiG and pre-processed with antiSMASH v.3 against five NRPs in the SMILES format (provided a single database tab-separated file)
nerpa.py -a test_data/MIBiG_subset/genome_predictions --smiles-tsv test_data/MIBiG_subset/structures_info.tsv
Example 3: a single reference genomes downloaded from NCBI RefSeq and pre-processed with antiSMASH v.5 against three NRP structures pre-processed with Nerpa (custom monomer graph format). Note: custom output directory is specified via -o; Nerpa algorithm for handling NRP-polyketide hybrids is enabled (recommended, uses rBAN).
nerpa.py -a test_data/NCBI_subset/genome_predictions_v5/ --structures test_data/NCBI_subset/structure.info.monomers -o NCBI_out_dir --process-hybrids
Check out the Nerpa main report (mind custom output directory path in Example 3):
less nerpa_results/latest/report.csv
For more details on the output directory content and its interpretation refer to the corresponding section.
To see the full list of available options, type
nerpa.py -h
All options are divided into four categories. The most important options in each category are listed below.
--process-hybrids process NRP-polyketide hybrid monomers (requires using rBAN)
--antismash-path <ANTISMASH_PATH> path to antiSMASH installation directory (should contain run_antismash.py); this option is required if genome sequences are provided in the FASTA or GenBank format and antiSMASH is not in PATH
--threads <THREADS> number of threads for running Nerpa
--output_dir <OUTPUT_DIR>, -o <OUTPUT_DIR> path to output dir [default: nerpa_results/results_<DATE_TIME>]; if not specified, a symlink nerpa_results/latest pointing to the output directory is created.
The most convenient way to get antiSMASH predictions of BGC in your genomic data with antiSMASH is to upload your
FASTA or GBK file to their webserver.
When the server job is completed, you may download archive with results ('Download -> Download all results'), unpack it and
provide the path to the unpacked directory or just the main JSON file from it to Nerpa via option -a.
You can also use the command-line version of antiSMASH.
Nerpa was tested with outputs from antiSMASH version 3.0, 5.0-5.2, 6.0. The recommended running parameters for v.5.1.1 are:
python run_antismash.py <your_genome.fasta> --output-dir <your_output_dir> --genefinding-tool prodigal --skip-zip-file --enable-nrps-pks --minimal
Note that you may specify an unlimited number of antiSMASH output files by using -a multiple times or by specifying a root directory with many inputs inside.
You may also write paths to all antiSMASH outputs in a single file and provide it via option --antismash_output_list.
TODO: describe they key options in this category. See the Nerpa help message for all options.
NRP molecules should be specified in the SMILES format.
One option is to provide them as a space-separated list of SMILES strings via option --smiles.
Another way is to write all structures in a multi-column file and specify it via --smiles-tsv.
Default column separator (\t), names of the SMILES column (SMILES) and the column with molecule IDs (row index) could be adjusted via option --sep, --col_smiles, and --col_id, respectively.
Alternatively, Nerpa-preprocessed structures could be specified in the custom text format via --structures. E.g., you may download the full preprocessed database of putative NRP structures here and provide it.
TODO: describe they key options in this category. See the Nerpa help message for all options.
--force-existing-outdir don't crash if the output directory already exists. Note: files in the output directory might be overwritten in this case!
The key files/directories inside the Nerpa output directory (see the --output_dir option) are:
reports.csvmatched NRP-BGC pairs with scoresdetailsdirectory with detailed descriptions and exact alignments for each match. Each file in the directory corresponds to an NRP and contains information on all matches with this NRP.
Currently, matches are not sorted by score, so please use GNU sort to get the sorted list:
cat <NERPA_OUTPUT_DIR>/report.csv | (read -r; printf "%s\n" "$REPLY"; sort -r -n -k1,1 -t ',') | less
Assume you have a genome and you want to predict NRP BGC(s) in it and check whether they might encode a known NRP. We will use Photobacterium galathea S2753 genome (NCBI GenBank ID: GCA_000695255.1) as an example; you can download it from NCBI in the command line by typing
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/695/255/GCA_000695255.1_Phalotolerans2753/GCA_000695255.1_Phalotolerans2753_genomic.fna.gz
gunzip GCA_000695255.1_Phalotolerans2753_genomic.fna.gz
Further you need to download the Nerpa-preprocessed pNRPdb, the database of putative NRP structures, from the Nerpa paper's zenodo entry:
wget https://zenodo.org/record/5503984/files/pnrpdb_preprocessed.info
See more detail on pNRPdb in the paper. To get the original structures in SMILES, get the raw database:
wget https://zenodo.org/record/5503984/files/pnrpdb_summary.tsv
If antiSMASH is installed in your system, just run Nerpa with the genome and the preprocessed database as
nerpa.py --sequences GCA_000695255.1_Phalotolerans2753_genomic.fna --structures pnrpdb_preprocessed.info --process-hybrids -o use_case_1_output
If you don't have antiSMASH installed, please process the genome using their web server (note: you can either upload your ungzipped FASTA/GenBank file or just specify NCBI ID). You can set 'Extra features: All off' or just rely on the default settings. Wait for the job completion and go to 'Download -> Download all results' or type
wget --no-check-certificate https://antismash.secondarymetabolites.org/upload/bacteria-<JOB_ID>/GCA_000695255.1_Phalotolerans2753_genomic.zip
unzip GCA_000695255.1_Phalotolerans2753_genomic.zip -d GCA_000695255.1_Phalotolerans2753_genomic
where <JOB_ID> is a unique antiSMASH job ID like df5ec71d-8f35-4787-b128-ea212e0e208a. After that, you can provide the unpacked antiSMASH output to Nerpa as
nerpa.py -a GCA_000695255.1_Phalotolerans2753_genomic --structures pnrpdb_preprocessed.info --process-hybrids -o use_case_1_output
Finally, check out the main results in use_case_1_output/report.csv (see also the Output files section). You should see the two highest scoring hits:
score,peptide,nrp len,match cnt,all matched,mol id,prediction id
12.3045,,0,6,0,NPA002859_variant0,./use_case_1_output/predictions/converted_antiSMASH_v5_outputs_ctg20_nrpspredictor2_codes_part0
12.3045,,0,6,0,NPA002702_variant0,./use_case_1_output/predictions/converted_antiSMASH_v5_outputs_ctg20_nrpspredictor2_codes_part0
These are ngercheumicin variants NPA002859 and NPA002702 from the NP Atlas database. The putative ngercheumicin is located in 20th contig of the P. galathea S2753 genome.
The Nerpa alignments for these hits are in use_case_1_output/details/NPA002859_variant0.match and use_case_1_output/details/NPA002702_variant0.match. E.g., type
cat use_case_1_output/details/NPA002702_variant0.match
to get all details
NPA002702_variant0
./use_case_1_output/predictions/converted_antiSMASH_v5_outputs_ctg20_nrpspredictor2_codes_part0
SCORE: 12.3045
ALIGNMENT:
ORF_ID A_domain_Idx Prediction_DL-config Prediction_Top_Residue Prediction_Top_Score Prediction_Modifications Matched_Residue Matched_Residue_Score Nerpa_Score Monomer_Idx Monomer_Code Monomer_DL-config Monomer_Residue Monomer_Modifications
ctg20_orf00048 0 D leu 100 - leu 100 1.86857 1 Leu NA leu -
ctg20_orf00048 1 L thr 100 - thr 100 2.73171 2 @L-aThr/Thr L thr -
ctg20_orf00048 2 D ser 100 - ser 100 3.60635 3 @D-Ser D ser -
ctg20_orf00048 3 L thr 100 - thr 100 2.73171 4 @L-aThr/Thr L thr -
ctg20_orf00048 4 D leu 100 - leu 100 1.86857 5 Leu NA leu -
ctg20_orf00048 5 L ile 80 - leu 70 -0.502364 6 Leu NA leu -
Assume you have an NRP and you want to check whether there a BGC producing this NRP is already sequence and available in NCBI. We will use ngercheumicin F (NP Atlas ID: NPA002702) as an example; SMILES of this compound is
CCCCCC/C=C\CCCC(CC(=O)NC(CC(C)C)C(=O)N[C@H]1[C@@H](OC(=O)C(NC(=O)C(NC(=O)[C@@H](NC(=O)[C@H](NC1=O)CO)[C@H](C)O)CC(C)C)CC(C)C)C)O
To look for a possible BGC of this compound in 13,399 reference and representative bacterial genomes from NCBI RefSeq, you can download their Nerpa-preprocessed genomic database available in the Nerpa paper's zenodo entry:
wget https://zenodo.org/record/5503984/files/bacterial_ref_and_repr_genomes_20210604_preprocessed.tar.gz
tar xzf bacterial_ref_and_repr_genomes_20210604_preprocessed.tar.gz --one-top-level
Next, run Nerpa with the compound and the preprocessed genomes as
nerpa.py -a bacterial_ref_and_repr_genomes_20210604_preprocessed --smiles "CCCCCC/C=C\CCCC(CC(=O)NC(CC(C)C)C(=O)N[C@H]1[C@@H](OC(=O)C(NC(=O)C(NC(=O)[C@@H](NC(=O)[C@H](NC1=O)CO)[C@H](C)O)CC(C)C)CC(C)C)C)O" --process-hybrids -o use_case_2_output
Note: mind quotation marks around the SMILES string!
Note 2: this run may be somewhat time-consuming since Nerpa compares the compound against 13,399 genomes, e.g., it may take about a day if you use a single thread.
Check out the main results in use_case_2_output/report.csv (see also the Output files section). You should see one highest scoring hit:
score,peptide,nrp len,match cnt,all matched,mol id,prediction id
12.3045,,0,6,0,compound_000000_variant0,/Bmo/agurevich/Nerpa/test_bioconda/use_case_2_output/predictions/GCA_000695255.1_Phalotolerans2753_genomic_ctg20_nrpspredictor2_codes_part0
5.70424,,0,5,0,compound_000000_variant0,/Bmo/agurevich/Nerpa/test_bioconda/use_case_2_output/predictions/GCA_003217475.1_ASM321747v1_genomic_ctg2_nrpspredictor2_codes_part0
We consider Nerpa scores below 6.0 as unreliable, so there is only one reasonable hit to a BGC predicted in GCA_000695255.1 (P. galathea S2753), a known ngercheumicin producer.
The corresponding Nerpa alignment is in use_case_2_output/details/compound_000000_variant0.match:
compound_000000_variant0
/Bmo/agurevich/Nerpa/test_bioconda/use_case_2_output/predictions/GCA_000695255.1_Phalotolerans2753_genomic_ctg20_nrpspredictor2_codes_part0
SCORE: 12.3045
ALIGNMENT:
ORF_ID A_domain_Idx Prediction_DL-config Prediction_Top_Residue Prediction_Top_Score Prediction_Modifications Matched_Residue Matched_Residue_Score Nerpa_Score Monomer_Idx Monomer_Code Monomer_DL-config Monomer_Residue Monomer_Modifications
ctg20_orf00048 0 D leu 100 - leu 100 1.86857 1 Leu NA leu -
ctg20_orf00048 1 L thr 100 - thr 100 2.73171 2 @L-aThr/Thr L thr -
ctg20_orf00048 2 D ser 100 - ser 100 3.60635 3 @D-Ser D ser -
ctg20_orf00048 3 L thr 100 - thr 100 2.73171 4 @L-aThr/Thr L thr -
ctg20_orf00048 4 D leu 100 - leu 100 1.86857 5 Leu NA leu -
ctg20_orf00048 5 L ile 80 - leu 70 -0.502364 6 Leu NA leu -
TODO: combine Use case 1 and Use case 2.
If you use Nerpa in your research, please cite Kunyavskaya, Tagirdzhanov et al., 2021. You may find the citation in various styles or download a bibtex record here.
You can leave your comments and bug reports at our GitHub repository tracker (recommended way) or sent it via e-mail: [email protected]
Your comments, bug reports, and suggestions are very welcomed. They will help us to improve Nerpa further. In particular, we would love to hear your thought on desired features of the future Nerpa web service (will be available from http:https://cab.cc.spbu.ru/ soon).
If you have any troubles running Nerpa, please attach nerpa.log from the directory <output_dir>.