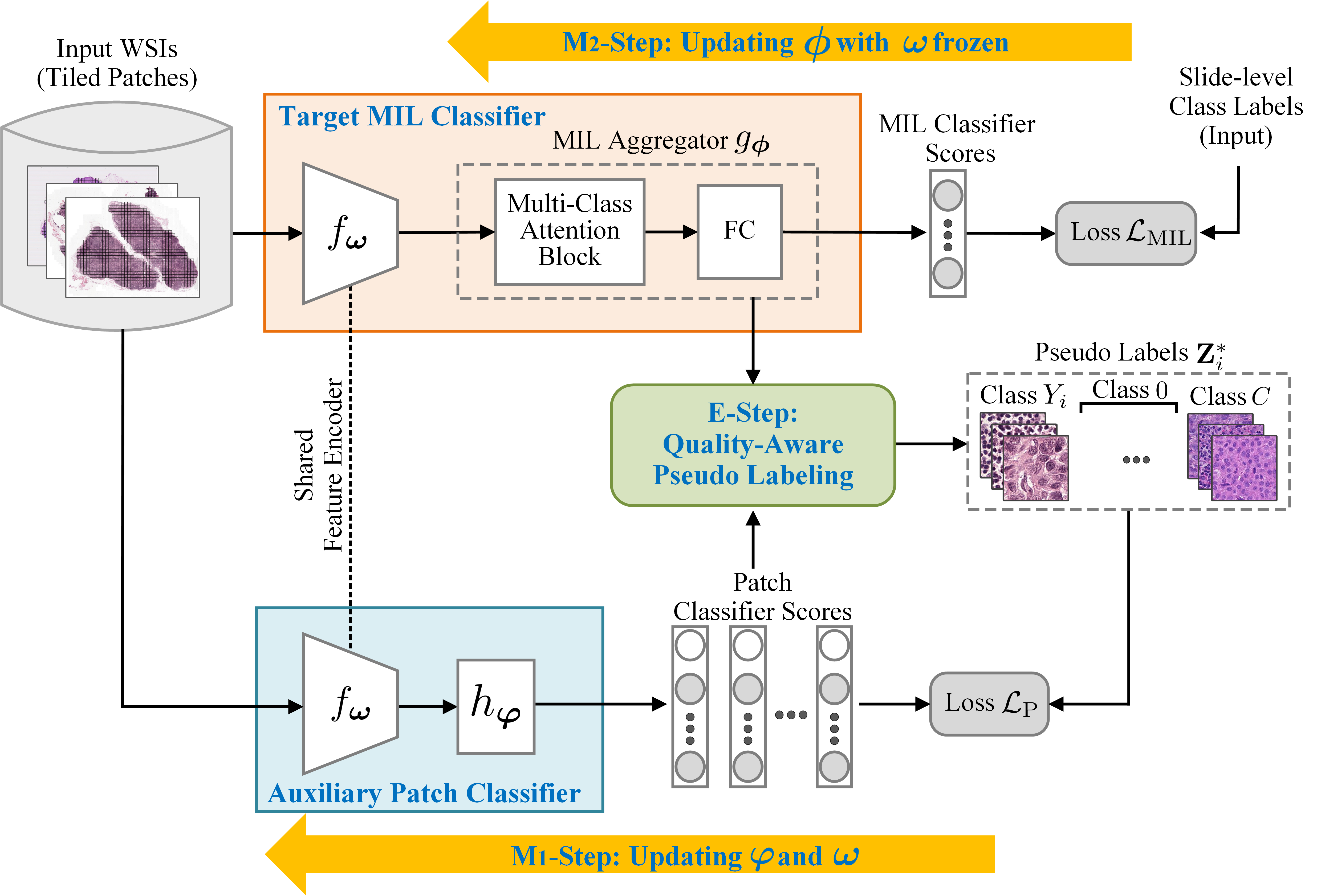
Whole-slide image (WSI) classification is fundamental to computational pathology, which is challenging in extra-high resolution, expensive manual annotation, data heterogeneity, etc. Multiple instance learning (MIL) provides a promising way to tackle these challenges. Nevertheless, due to the gigapixel resolutions, MIL for WSI classification inherently suffers from the memory bottleneck issue, which remains largely unresolved in the literature. Towards this end, this paper presents a Bayesian Collaborative Learning (BCL) framework for gigapixel WSI classification. The basic idea of BCL is to introduce an auxiliary patch classifier, along with the target MIL classifier to be learned, so that the two classifiers can collaborate with each other to achieve joint learning of the feature encoder and the MIL aggregator in the MIL classifier, free of the memory bottleneck issue. Such a collaborative learning procedure is formulated under a principled Bayesian probabilistic framework and an Expectation-Maximization algorithm is developed to infer the optimal model parameters iteratively. As an implementation of the E-step, an effective quality- aware pseudo labeling approach is further proposed. Our BCL is extensively evaluated on three publicly available WSI datasets, i.e., CAMELYON16, TCGA-NSCLC and TCGA-RCC, achieving an AUC of 95.6%, 96.0% and 97.5% respectively, which consistently outperforms all the compared MIL methods. Comprehensive analysis and discussion will also be presented for in-depth understanding of the proposed BCL.
The trained model weights and are provided here (Google Drive | Baidu Cloud (bclm)). You can download these files and drag bcl_model.pth and to the result/CAMELYON directory.
-
Camelyon16
Camelyon16 is a public challenge dataset of sentinel lymph node biopsy of early-stage breast cancer, which includes 270 H&E-stained WSIs for training and 129 for testing (48 LNM-positive and 81 LNM-negative), collected from two medical centers.
Download from here. -
TCGA
TCGA-NSCLC and TCGA-RCC are public available datasets for non-small-cell lung carcinoma(NSCLC) subtyping and renal cell carcinoma(RCC) subtyping respectively. They can be download from here
Datasets are preprocessed according to CLAM, which extracts 256x256 patches from each WSI and converts them into 1024-dimensional feature vectors. All patch features are saved in .pt format, and coordinates of each patch are also stored in the directory.
The feature encoder is initialized by using ResNet50 backbone pretrained on ImageNet, the MIL classifier is initialized by training Attention-based MIL, which provides attention weight of each patch for pseudo labeling and start iterations. So, you can initialize the BCL model by setting args.round=0 and running the following command:
python M2_update_MIL_classifier.py --round 0 --results_dir result/CAMELYON
Then initialized model and attention weights will be stored in result/CAMELYON directory, you can continue to assign pseudo labels and start EM iterations by following:
python E_pseudo_labeling.py --round 1 --results_dir result/CAMELYON
You can retrain the feature extractor and patch classifier with pseudo labels:
python M1_update_feat_encoder.py --round 1 --results_dir result/CAMELYON
After all above, you have updated the patch-level feature extractor once. By running extract_feature_clean.py, all patch features will be updated. So, you can retrained the MIL classifier with updated patch features:
python M2_update_MIL_classifier.py --results_dir result/CAMELYON --round 1
Until this step, one round of EM iteration has completed. Continue to iterate until the model converges.
First, you should drag pretrained weights of the feature encoder t4_feature_extractor.pth to the result/CAMELYON directory, and extract the patch features by running extract_feature_clean.py with that.
Then you can test the performance of BCL on Camelyon16 dataset by following code after draging the pretrained weights of model bcl_model.pth to the result/CAMELYON directory:
python M2_update_MIL_classifier.py --is_test --load_model
The key to BCL is to introduce an auxiliary patch classifier to improve the MIL classifier collaboratively. Here we further visualize some representative results to show this point intuitively. More precisely, we choose two exemplary WSIs (named “test 021” and “test 122”) from CAMEL YON16 for visualization. We depict the MIL-classifier heat maps obtained by BCL at the first and last iteration steps and also by other methods and the patch-classifier heat maps obtained by BCL at various iteration steps. For the learned patch classifier, we also visualize its Class Activation Maps (CAMs) generated over some patches selected from these two WSIs.
If you find our work useful in your research or if you use parts of this code please consider citing our paper.