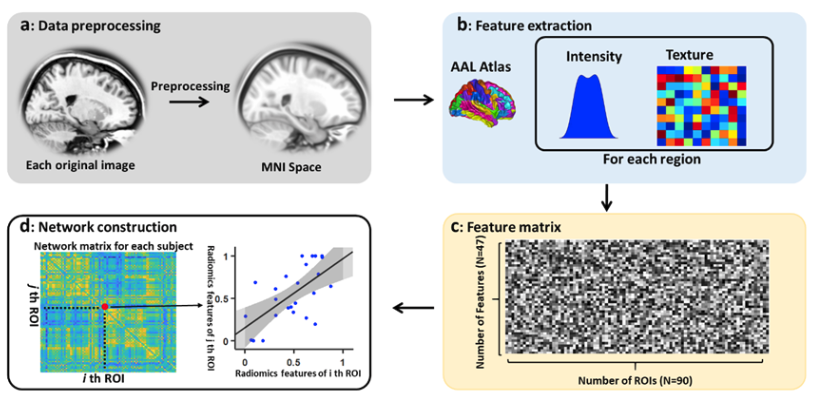
R2SNs provide a novel, reliable and biologically plausible method to understand human morphological covariance based on sMRI.
If you use R2SN_code, please cite this orignal paper: Original Paper
Zhao K, Zheng Q, Che T, et al. Regional radiomics similarity networks (R2SNs) in the human brain: Reproducibility, small-world properties and a biological basis [J]. Network Neuroscience, 2021, 1-15.
Install option 1 (for installing the R2SN_construction code in a chosen directory): clone repository, install locally
-
Navigate to the main R2SN_construction directory , then run:
pip install .
pip install R2SN
- Python >= 3.8
- Nibabel >= 3.2.1
- Pyradiomics >= 3.0.1
- Scipy >= 1.6.2
- SimpleITK >= 2.1.1.2
- NumPy >= 1.21.5
- Pandas >= 1.2.3
- Sklearn
from R2SN import feature_extraction
if __name__ == '__main__':
Image_path = r'D:\Python_project\venv\Brain\R2SN\R2SN\data\Image' # input image path
Network_output_path = r'D:\Python_project\venv\Brain\R2SN\R2SN\data\Network' # output network path
n_jobs = 1 # number of process
feature_extraction.R2SN_feature_extract(Image_path = Image_path,
Network_output_path = Network_output_path,
n_jobs = n_jobs)
Here, the image_path is the dir of your original file. The Network_out_path is the dir of your choosed ouput dir. n_jobs is means the number of jobs runing simultaneously.
Author: Fan Yang ([email protected]),Kun Zhao ([email protected])
Any questions, pls do not hesitate to contact [email protected]
The registeration section is based on ANTs toolkit.