Project Page | Video | Paper
This repository is an unofficial implementation of the paper Learning to Reconstruct Botanical Trees from Single Images.
I recommend using conda environments for managing dependencies. I have provided a conda environment setup file which provides all the dependencies needed for the project. Create a conda environment by running:
conda create --name <env> --file requirements.txtThe paper uses three different neural networks. One for segmenting leaves and branches, one for predicting the Radial Bounding Volume (RBV) and one for predicting the species of the tree. The repository is organized in the following directory structure:
botanical-trees
├── dataset code for data generation
├── rbv_prediction code for training RBV prediction network
├── scripts SLURM scripts for training, augmentation, etc.
├── segmentation code for training the segmentation network
└── utils scripts for data processing and augmentation
Note: Code for training the species classification network is not present.
I have used Blender along with the Modular Tree to generate the dataset. The dataset includes realistic renderings of the tree, corresponding segmentation mask and the RBV. To generate your own version of the dataset use the tree_gen.py script. Presets for generating different species of trees are present in dataset/presets directory.
To train the segmentation network, run:
python segmentation/main.py <path_to_dataset> <path_to_results> --epochs 25 --batch_size 2 --num_classes 3 --encoder 'tu-xception41'To train the RBV prediction network on ground truth segmentation mask, run:
python rbv_prediction/main.py <path_to_dataset> --use_gt --gpus=1 --batch_size=32 --lr 0.001Note: All experiments are tracked with Weights and Biases. Be sure to change the project location in main.py files for both networks or if you don't want experiment tracking disable WandB by running wandb off.
Feel free to raise GitHub issues if you find anything concerning. Pull requests adding additional features are welcome too.
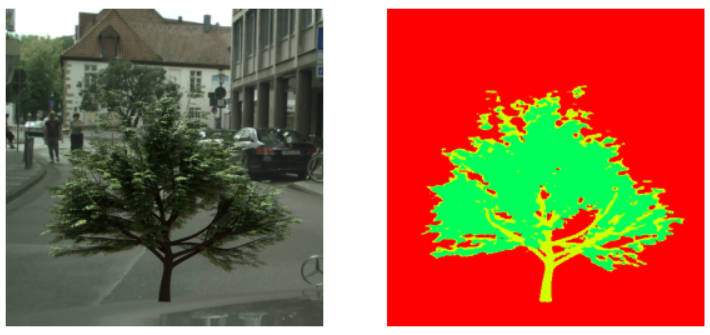
The RBV prediction network is successful in predicting the overall shape of the tree however it can’t predict radii of sectors in a single layer correctly as seen in the results above. I could not figure out why this is happening so if you do, please open a PR 😜.
If you find this code useful in your research then please cite the original authors.
@article{10.1145/3478513.3480525,
author = {Li, Bosheng and Ka\l{}u\.{z}ny, Jacek and Klein, Jonathan and Michels, Dominik L. and Pa\l{}ubicki, Wojtek and Benes, Bedrich and Pirk, S\"{o}ren},
title = {Learning to Reconstruct Botanical Trees from Single Images},
year = {2021},
url = {https://doi.org/10.1145/3478513.3480525},
doi = {10.1145/3478513.3480525},
journal = {ACM Trans. Graph.},
}