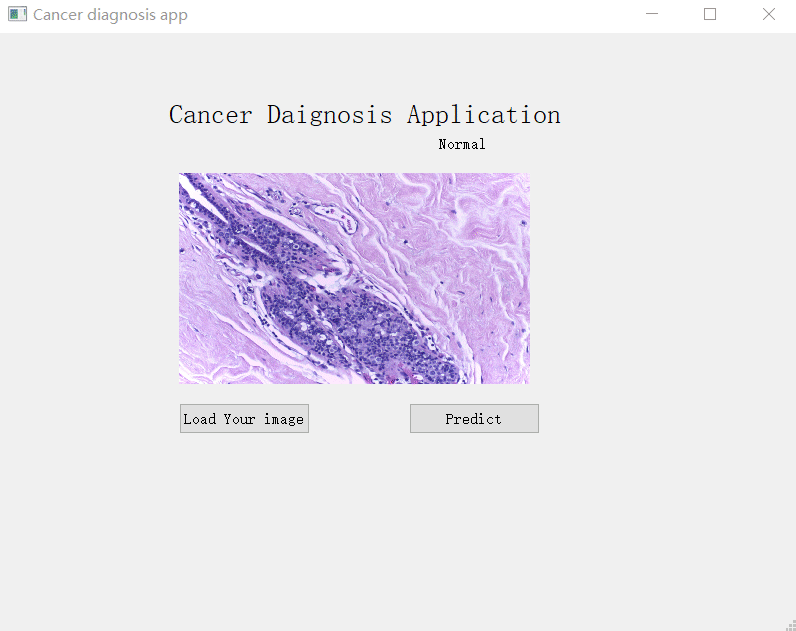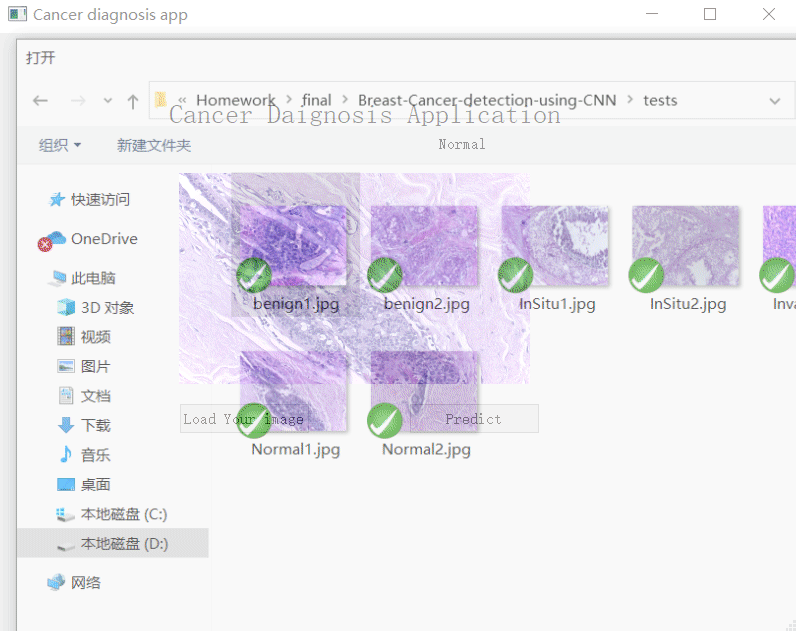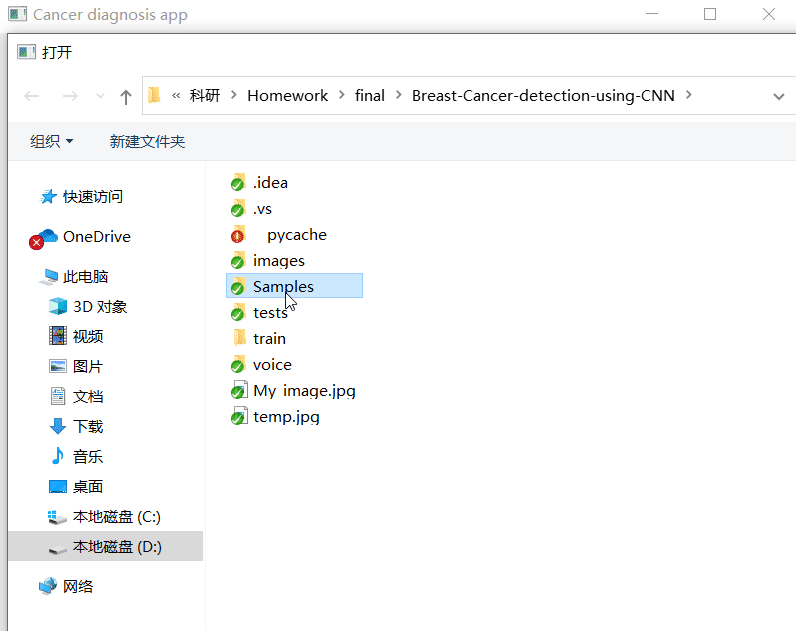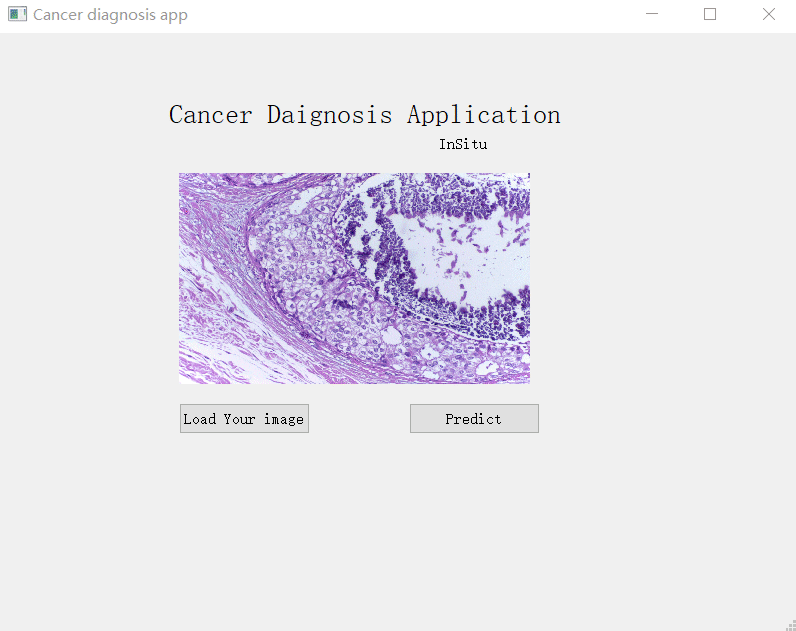
Utilised transfer learning to fine-tune a CNN model trained on BACH on TUPAC dataset to build breast cancer subtype classifier and did a simple windows Qt UI for it, users can click to upload a breast HP image, and the app will tell you which type it is, specifically Benign, Normal, Invasive, In Situ.
Breast cancer represents the second mortality among cancers in women today. There are many techniques to diagnose breast cancer, among which the pathological diagnosis is regarded as the golden standard. At present, there are studies about computer-aided diagnosis systems to reduce the cost and increase the efficiency of this process. Traditionally, previous work usually is based on feature-engineering, which is labor-intensive and tedious. To alleviate these challenges, we use deep learning methods to build a Computer Aided System. Compared with feature-based methods, a method for the classification of hematology and eosin-stained breast biopsy images using Convolutional Neural Networks (CNNs) is proposed. CNNs has been a quick and accurate in image classification tasks. We aim to help humans detect breast cancer by filtering out normal cases with well-trained CNN, thereby reducing doctors’ unnecessary work. In this paper, we train a CNN to evaluate/detect breast cancer and reduce unnecessary testing. Specially, Images are classified into four classes, normal tissue, benign lesion, in situ carcinoma, and invasive carcinoma, and in two classes, carcinoma and non-carcinoma. The proposed model is designed to detect BrC by retrieving features at different scales, including fine-grained scale (nuclei) and coarse-grained scale (overall tissue organization). We conduct experiment on BACH dataset, and achieve good performance, which is 77.8% on accuracy and the sensitivity of our method for cancer cases is 95.6%. This design allows the extension of the proposed system to whole-slide histology images. Accuracies of 77.8% for four classes is achieved. The sensitivity of our method for cancer cases is 95.6%. Dataset:in the preprocessed npy files
 Examples of microscopic biopsy images in the dataset: (A) normal; (B) benign; (C) in situ carcinoma; and (D) invasive carcinoma
Examples of microscopic biopsy images in the dataset: (A) normal; (B) benign; (C) in situ carcinoma; and (D) invasive carcinoma
We use a pre-trained CNN model on Keras to predict the subtype of breast cancer.
You can download all the files and run main.py and it will generate a UI interface in which you can upload images and get a prediction.
Or you can also use the console to train a model or make predictions for a bunch of images in a file.
Dataset:in the preprocessed npy files.
https://drive.google.com/drive/folders/17LR9ssbENit-3vsEAM63FptNasB5AHrr
- Collected data from open-source datasets(TUPAC) for breast cancer images
- Crop and resized the images into the same size(512*384)
- Used data augmentation techniques (rotate, shift, and flip images randomly) to promote its generalisation ability
- Trained the CNN for image classification to divide those images into four types, which are "invasive", "benign", "normal", "in Situ". And achieved an average accuracy "of 77%" for classifying cancer images.
Used a python package tts-wrapper to turn the diagnosis result text into voice.
My network has 21 layers, the train data comes from the npy files on Google cloud disk, which is pre-processed and can grab up to use. My train data set has 4776 cases, and my evaluation data set has 240 cases. My batch size is 16, after training 20 epochs, my model can already have a 77% accuracy and 60% percent specificity. If you don't want to train a new model, you can still make predictions. Since I uploaded my model(my_model.h5)
You can choose a whole-slide histology image and make predictions using the model. The result will be the probability of each section.
I use PyQt4 to do the simple UI interface which has a button to upload an image, and another button to predict the result.
I use the Aliyun API to generate 4 sound files from text and recorded them. It will play one of the pre-recorded files after diagnosing one type of breast cancer.