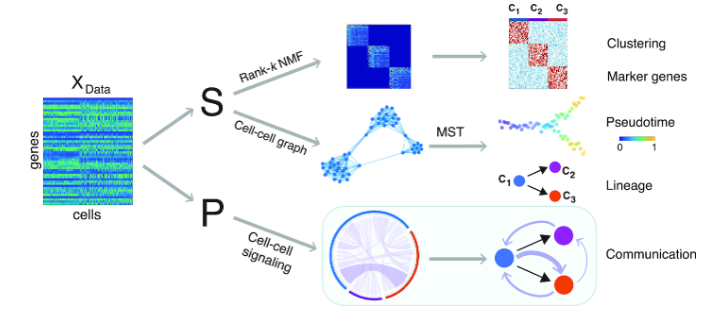
Package used for the inference of clustering, cell lineage, pseudotime and cell-cell communication network from scRNA-seq data.
- SoptSC is also avaiable as a R package at: https://mkarikom.github.io/RSoptSC/
> 1. Estimation of the number of clusters from the single-cell data.
> 2. Identification of cell subpopulations.
> 3. Identification of DEGs for each cell subpopulation.
> 4. Inference of pseudotime in an unsupervised manner: i.e., initial cell is not required.
> 5. Inference of cell lineage and trajectory in an unsupervised manner: i.e., initial cluster is not required.
> 6. Inference of cell-cell communication network given a group of Ligand-Receptor pairs and their downstream target genes.
- Requires: MATLAB R2019 or later.
- Click here to go through all the steps
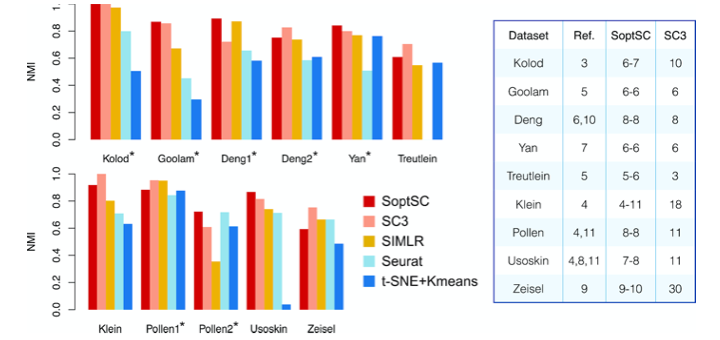
- Data used for each figure is deposit under folder 'Data'
- Please refer to each M file for more detailed descriptions of the corresponding function.
- Cell lineage and communication network inference via optimization for single-cell transcriptomics, S. Wang, et al., Nucleic acids research, 2019.
- Data used for this tutorial comes from Single cell transcriptomics of human epidermis identifies basal stem cell transition states, S. Wang, et al.,Nature Communications, 2020.
Please feel free to contact us if you have any question: shuxionw 'at' uci 'dot' edu