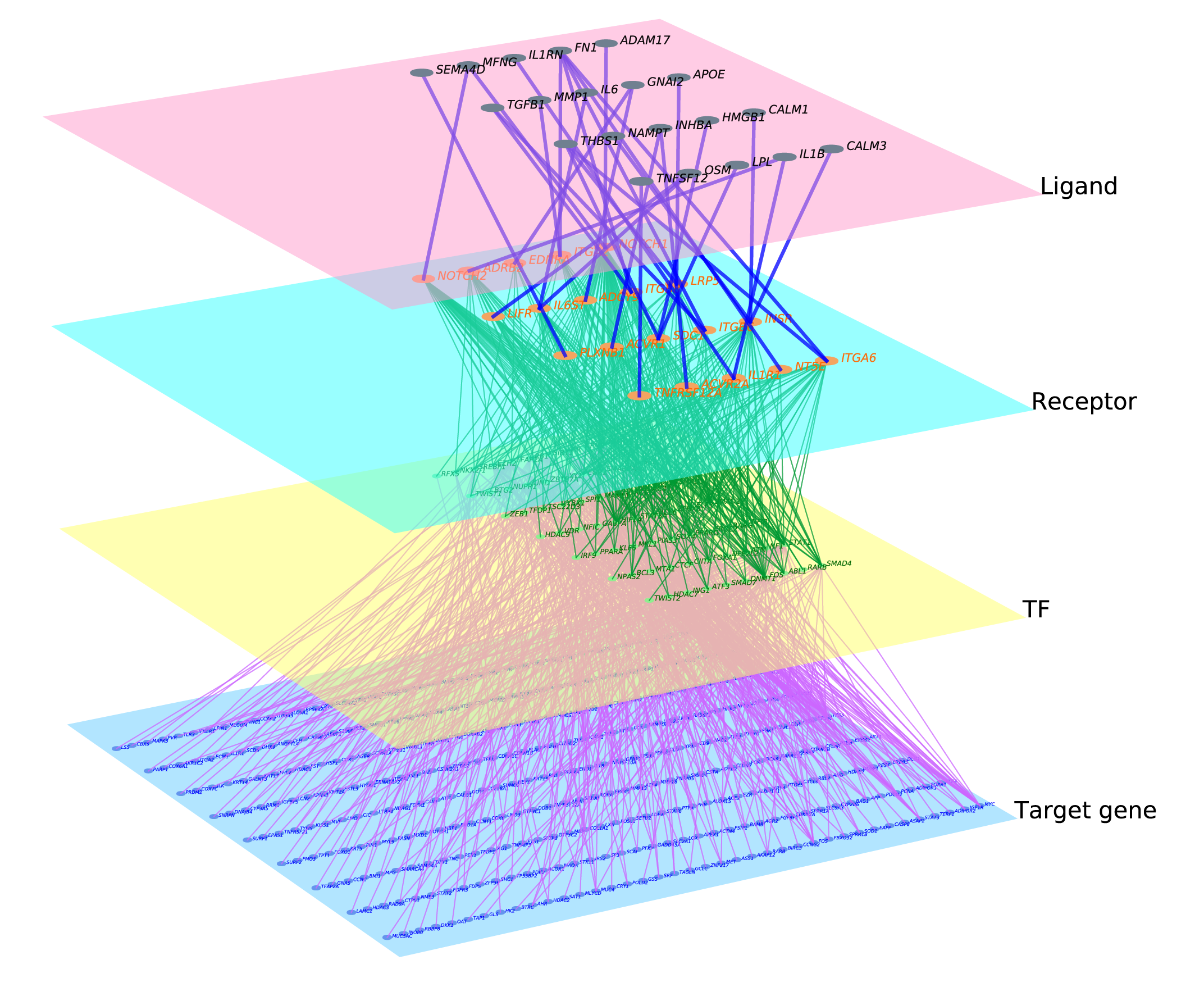
scMLnet is an R package developed to construct inter-/intracellular multilayer singaling network based on single-cell RNA-seq expression data. scMLnet constructs the multilayer network by integrating intercellular pathways (ligand-receptor interactions) and intracellular subnetworks (receptor-TF pathways and TF-target gene interactions) based on cell-type specific gene expression, prior network information and statistical inference. scMLnet can also visualize the constructed inter-/intracellular signaling pathways between the central cell and neighboring cells. scMLnet is implemented using R (version 3.6.0) and Python (version 3.7).
The main steps of the scMLnet algorithm include:
- Step1 Constructing Ligand-Receptor subnetwork: defines potential Ligand-Receptor subnetworks from scRNA-Seq data and Ligand-Receptor database by getting highly expressed genes (HEGs). HEGs in Type A (sender cells) are considered as potential ligands and HEGs in Type B (receiver cells) as potential receptors.
- Step2 Constructing TF-Target gene subnetwork: defines potient TF-Target gene subnetworks from scRNA-Seq data and TF-Target gene database by getting HEG and Fisher’s exact test. HEGs in Type B are considered as potential target genes. Activated TFs can be inferred from the TF-Target gene subnetwork.
- Step3 Constructing Receptor-TF subnetwork: defines potient Receptor-TF subnetworks from activated TFs and Receptor-TF database by Fisher’s exact test. Activated receptors can be inferred from the TF-Target gene subnetwork.
- Step4 constructing multi-layer signaling network: defines multi-layer signaling network by performing correlation analysis between receptors and TFs, TFs and target genes and then overlapping the Ligand-Receptor, TF-Target gene, Receptor-TF subnetworks according to common receptors and TFs.
The python package pymnet module should be installed for visualizing the multi-layer singal network, you can download the source files directly from bitbucket or github, unzip and install:
python setup.py install
and then copy the /pymnet/sampling folder to the installation path (%PYTHONHOME%/Lib/site-packages/pymnet-0.1-py3.7.egg/pymnet). Alternatively, you can simply copy the /pymnet/ folder to your python library path (%PYTHONHOME%/Lib/site-packages). NOTE: The following python packages should be installed first: networkx, matplotlib.
The following R packages should be installed for creating the multi-layer singal network, please install before the installation of scMLnet:
- Seurat
- parallel
install scMLnet from github:
install.packages("devtools")
library(devtools)
devtools::install_github("YUZIXD/scMLnet")
library(scMLnet)
or install scMLnet module from the source code:
install.packages("path/to/download/scMLnet_0.1.0.tar.gz", repos = NULL, type = "source")
library(scMLnet)
Before using scMLnet, scRNA-seq data should be processed and clustered to identify cell types for dissecting cell type-specific gene expressions by employing existing methods or tools (e.g., Seurat). scMLnet requires the following information as input:
(1) scRNA-Seq expression matrix (a Sparse matrix, where rows represent genes, columns represent cells);
(2) clustering results containing two columns: cell’s barcode and cluster identities;
(3) two cluster identities of sending cells and receiving cells.
The output of scMLnet has two forms:
(1) tabular information of the constructed multilayer network, containing gene pairs connecting each upstream layer and downstream layer (i.e., Ligand_Receptor, Receptor_TF and TF_Gene subnetworks);
(2) visualization of the constructed multilayer network using the pymnet library.
The Working Directory requires the following files and directories:
| File | Description |
|---|---|
| /example | Input directory including scRNA-Seq data and clustering results |
| /database | Prior information about interactions between ligands, receptors, TFs and target genes |
| /database/LigRec.txt | The Ligand-Receptor interactions including three colums: Ligand, Receptor and Key (links connecting ligands with receptors by underlined) |
| /database/RecTF.txt | The Receptor-TF interactions including three colums: Receptor, TF and Key (links connecting receptors with TFs by underlined) |
| /database/TFTargetGene.txt | The TF-Target gene interactions including three colums: TF, Target gene and Key (links connecting TFs with Target genes by underlined) |
| /code/DrawNetNew.py | The python script for visualization |
| /output | Output directory, tabular and graphic results of the multi-layer signaling network will be saved in this folder |
A demonstration of using scMLnet to construct the multi-layer signaling network between B cells and Secretory cells from scRNA-Seq data of COVID-19 patients BALF can be found at following vignette. The expression matrix and annotation of clstuers can be found in the /example folder and the prior information about interactions in the /database folder.
- Vignette: Tutorial of scMLnet
Please cite the following paper when you use scMLnet:
Cheng J, Zhang J, Wu Z, Sun X*. Inferring microenvironmental regulation of gene expression from single-cell RNA sequencing data using scMLnet with an application to COVID-19. Briefings in Bioinformatics. 2020. Bbaa327.
The R code used for analysis in this paper is available at https://github.com/SunXQlab/paper-covid19